Abstract
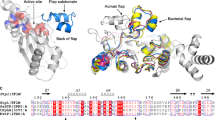
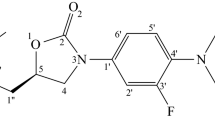
YsxC from Staphylococcus aureus is a member of the GTPase protein family, and is involved in the ribosomal assembly and stability of this microorganism through its interactions with the L17, S2 and S10 ribosomal proteins. Inhibition of its interactions with L17, S2, S10 and the β′ subunit of RNA polymerase influences ribosomal assembly, which may affect the growth of the microorganism. This makes YsxC a novel target for the design of inhibitors to treat the disease caused by S. aureus. Understanding the interaction mechanism between YsxC and its partners would aid in the identification of potential catalytic residues, which could then be targeted to inhibit its function. Accordingly, in the present study, an in silico analysis of the interactions between YsxC and L17, S2 and S10 was performed, and the potential residues involved in these interactions were identified. Based on the simulation results, a possible mechanism for the interactions between these proteins was also proposed. Finally, six ligands from among a library of 81,000 chemical molecules were found to interact with parts of the G2 and switch II regions of the YsxC protein. Moreover, their interactions with the YsxC protein were observed to provoke changes at its GTP-binding site, which suggests that the binding of these ligands leads to a reduction in GTPase activity, and they were also found to affect the interactions of YsxC with its partners. This observation indicates that the proposed interacting site of YsxC may act as an allosteric site, and disrupting interactions at this site might lead to novel allosteric inhibition of the YsxC protein.












Similar content being viewed by others
References
Chambers HF (2005) Community-associated MRSA-resistance and virulence converge. N Engl J Med 352:1485–1487. doi:10.1056/NEJMe058023
NABI Biopharmaceuticals (2007) Key facts about S. aureus infections (2007) NABI Biopharmaceuticals, Miami. http://www.nabi.com. Accessed 10 April 2007
Lowy FD (1998) Staphylococcus aureus infections. N Engl J Med 339:520–530. doi:10.1056/NEJM199808203390806
Lindsay JA, Holden MT (2006) Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Func Integr Genomics 6:186–201. doi:10.1007/s10142-005-0019-7
Tejerina C, Reig A, Codina J, Safont J, Baena P, Mirabet V (1992) An epidemiological study of burn patients hospitalized in Valencia, Spain during 1989. Burns 18:15–18. doi:10.1016/0305-4179(92)90112-8
Centers for Disease Control and Prevention (2003) Community-associated MRSA: frequently asked questions. Centers for Disease Control and Prevention, Atlanta. http://www.cdc.gov/ncidod. Accessed Aug 2003
Centers for Disease Control and Prevention. Community-associated MRSA: fact sheet (2003) Centers for Disease Control and Prevention, Atlanta. http://www.cdc.gov/ncidod. Accessed Feb 2003
Shopsin B, Mathema B, Martinez J, Ha E, Campo ML, Fierman A, Krasinski K, Kornblum J, Alcabes P, Waddington M et al (2000) Prevalence of methicillin-resistant and methicillin-susceptible Staphylococcus aureus in the community. J Infect Dis 182:359–362. doi:10.1086/315695
Keiichi H (2001) Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect Dis 1:147–155. doi:10.1016/S1473-3099(01)00091-3
Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC (1999) Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165–2169. doi:10.1126/science.286.5447.2165
Schaefer L, Uicker WC, Wicker-Planquart C, Foucher AE, Jault JM, Britton RA (2006) Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis. J Bacteriol 188:8252–8258. doi:10.1128/JB.01213-06
Cooper EL, García-Lara J, Foster SJ (2009) YsxC, an essential protein in Staphylococcus aureus crucial for ribosome assembly/stability. BMC Microbiol 9:266. doi:10.1186/1471-2180-9-266
Prágai Z, Harwood CR (2000) YsxC, a putative GTP-binding protein essential for growth of Bacillus subtilis 168. J Bacteriol 182:6819–6823
Wicker-Planquart C, Foucher AE, Louwagie M, Britton RA, Jault JM (2008) Interactions of an essential Bacillus subtilis GTPase, YsxC, with ribosomes. J Bacteriol 190:681–690. doi:10.1128/JB.01193-07
Christopoulos A, May LT, Avlani VA, Sexton PM (2004) G-protein-coupled receptor allosterism: the promise and the problem(s). Biochem Soc Trans 32:873–877. doi:10.1042/BST0320873
May LT, Leach K, Sexton PM, Christopoulos A (2007) Allosteric modulation of G protein-coupled receptors. Annu Rev Pharmacol Toxicol 47:1–51. doi:10.1146/annurev.pharmtox.47.120505.105159
Ruzheinikov SN, Das SK, Sedelnikova SE, Baker PJ, Artymiuk PJ, Garcia-Lara J, Foster SJ, Rice DW (2004) Analysis of the open and closed conformations of the GTP-binding protein YsxC from Bacillus subtilis. J Mol Biol 339:265–278. doi:10.1016/j.jmb.2004.03.043
Vassylyev DG, Shirouzu M, Wada T, Yokoyama S (2009) The crystal structure of the bacteria-specific L17 ribosomal protein from Thermus thermophilus. doi:10.2210/pdb1gd8/pdb
Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V (2000) Functional insights from the structure of the 30 S ribosomal subunit and its interactions with antibiotics. Nature 407:340–348. doi:10.1038/35030019
Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, van Roey P, Agrawal RK, Harvey ST, Sali A, Chapman MS, Frank J (2003) Study of the structural dynamics of the E. coli 70 S ribosome using real-space refinement. Cell 113:789–801. doi:10.1016/S0092-8674(03)00427-6
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Eswar N, Webb B, Marti-Renom MA, Madhusudhan M, Eramian D, Shen MY, Pieper U, Sali A (2007) Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci 50:2.9.1–2.9.31. doi:10.1002/0471140864.ps0209s50
Eramian D, Shen MY, Devos D, Melo F, Sali A, Marti-Renom MA (2006) A composite score for predicting errors in protein structure models. Protein Sci 15:1653–1666. doi:10.1110/ps.062095806
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theor Comput 4:435–447. doi:10.1021/ct700301q
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236. doi:10.1021/ja9621760
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935. doi:10.1063/1.445869
Kawata M, Nagashima U (2001) Particle mesh Ewald method for three-dimensional systems with two-dimensional periodicity. Chem Phys Lett 340:165–172. doi:10.1016/S0009-2614(01)00393-1
Hess B, Bekker H, Fraaije J, Berendsen HJC (1997) A linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472. doi:10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H
Miyamoto S, Kollman PA (1992) SETTLE—an analytical version of the shake and rattle algorithm for rigid water models. J Comput Chem 13:952–962. doi:10.1002/jcc.540130805
Martonák R, Laio A, Parrinello M (2003) Predicting crystal structures: the Parrinello–Rahman method revisited. Phys Rev Lett 90:075503. doi:10.1103/PhysRevLett.90.075503
Laskoswki RA, MacArthur MW, Moss DS, Thorton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi:10.1107/S0021889892009944
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519. doi:10.1002/pro.5560020916
Luthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356:83–85. doi:10.1038/356083a0
Brylinski M, Skolnick J (2008) A threading-based method (FINDSITE) for ligand binding site prediction and functional annotation. Proc Natl Acad Sci USA 105:129–134. doi:10.1073/pnas.0707684105
Laurie AT, Jackson RM (2005) Q-SiteFinder: an energy-based method for the prediction of protein–ligand binding sites. Bioinformatics 21:1908–1916. doi:10.1093/bioinformatics/bti315
Pagni M, Ioannidis V, Cerutti L, Zahn-Zabal M, Jongeneel CV, Hau J, Martin O, Kuznetsov D, Falquet L (2007) MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res 35:W433–W437. doi:10.1093/nar/gkm352
Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH (2009) CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res 37(D):205–210. doi:10.1093/nar/gkn845
Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32(W):327–331. doi:10.1093/nar/gkh454
von Grotthuss M, Pas J, Rychlewski L (2003) Ligand-Info, searching for similar small compounds using index profiles. Bioinformatics 19:1041–1042. doi:10.1093/bioinformatics/btg117
Schuettelkopf AW, van Aalten DMF (2004) PRODRG—a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr D60:1355–1363. doi:10.1107/S0907444904011679
de Vries SJ, van Dijk ADJ, Bonvin AM (2006) WHISCY: what information does surface conservation yield? Application to data-driven docking. Protein Struct Funct Bioinf 63:479–489. doi:10.1002/prot.20842
Chen H, Zhou HX (2005) Prediction of interface residues in protein-protein complexes by a consensus neural network method: test against NMR data. Protein Struct Funct Bioinf 61:21–35. doi:10.1002/prot.20514
Zhou HX, Shan Y (2001) Prediction of protein interaction sites from sequence profiles and residue neighbor list. Protein Struct Funct Bioinf 44:336–343. doi:10.1002/prot.1099
Duhovny D, Nussinov R, Wolfson HJ (2002) Efficient unbound docking of rigid molecules. Lect Notes Comput Sci 2452:185–200. doi:10.1007/3-540-45784-4_14
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33:W363–W367. doi:10.1093/nar/gki481
Andrusier N, Nussinov R, Wolfson HJ (2007) FireDock: fast interaction refinement in molecular docking. Protein Struct Funct Bioinf 69:139–159. doi:10.1002/prot.21495
Dominguez C, Boelens R, Bonvin AM (2003) HADDOCK: a protein–protein docking approach based on biochemical and/or biophysical information. J Am Chem Soc 125:1731–1737. doi:10.1021/ja026939x
de Vries SJ, van Dijk AD, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AM (2007) HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Protein Struct Funct Bioinf 69:726–733. doi:10.1002/prot.21723
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng 8:127–134. doi:10.1093/protein/8.2.127
Lefèvre F, Rémy MH, Masson JM (1997) Alanine-stretch scanning mutagenesis: a simple and efficient method to probe protein structure and function. Nucleic Acids Res 25:447–448. doi:10.1093/nar/25.2.447
Feyfant E, Sali A, Fiser A (2007) Modeling mutations in protein structures. Protein Sci 16:2030–2041. doi:10.1110/ps.072855507
Ditlev EB, William MCJ, Andrew PC, Brian TW, Ramakrishnan V (2002) Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol 316:725–768. doi:10.1006/jmbi.2001.5359
Acknowledgments
RK, Manivel and Kannan thank the University Grants Commission, Government of India (UGC) for funding this Major Research Project, and the Rajiv Gandhi National Fellowship for also providing funding assistance. Research at the Laboratory of the Centre for Excellence in Bioinformatics, Pondicherry University, India is funded by the Department of Information Technology (DIT) and the Department of Biotechnology (DBT), Government of India, New Delhi, India.
Author information
Authors and Affiliations
Corresponding author
Additional information
Amit Goyal and Muthu Kannan contributed equally to this work.
Rights and permissions
About this article
Cite this article
Goyal, A., Muthu, K., Panneerselvam, M. et al. Molecular dynamics simulation of the Staphylococcus aureus YsxC protein: molecular insights into ribosome assembly and allosteric inhibition of the protein. J Mol Model 17, 3129–3149 (2011). https://doi.org/10.1007/s00894-011-0998-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-011-0998-3