Abstract
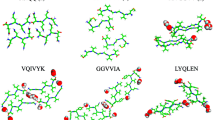
The aggregation modes of hexapeptide fragments of Tau, Insulin and Aβ peptide (VQIVYK, MVGGVV and LYQLEN) were found from their microcrystalline structures that had been recently resolved by X-ray analysis. The atomic structures reveal a dry self-complementary interface between the neighboring β-sheet layers, termed “steric zipper”. In this study we perform several all-atom molecular dynamics simulations with explicit water to analyze stability of the crystalline fragments of 2-10 hexapeptides each and their analogs with single glycine replacement mutations to investigate the structural stability, aggregation behavior and thermodynamic of the amyloid oligomers. Upon comparing single and double layer models, our results reveal that additional strands contribute significantly to the structural stability of the peptide oligomers for double layer model, while in the case of single layer model the stability decreases (or remains the same in the case of LYQLEN). This is in agreement with the previous studies performed on different types of amyloid models. We also replaced the side-chains participating in the steric zipper interfaces with glycine. None of the mutants were structurally stable compared to the respective wild type model, except for mutants V2G and V6G in MVGGVV2 case. The exception can be explained by structural features of this particular polymorph. The double layer decamer and dodecamer aggregates of the wild type hexapeptides appear to be stable at 300K, which is confirmed by the conservation of high anti-parallel β-sheet content throughout the whole simulation time. Deletions of the side chains resulted in decline of secondary structure content compared to corresponding wild type indicating that the role of the replaced amino acid in stabilizing the structure. Detailed analysis of the binding energy reveals that stability of these peptide aggregates is determined mainly by the van der Waals and hydrophobic forces that can serve as quantitative measure of shape complementarities between the side chains. This observation implies that interactions among side chains forming the dehydrated steric zipper, rather than among those exposed to water, are the major structural determinant. The electrostatic repulsion destabilizes the studied double layer aggregates in two cases, while stabilizes the other two. Negative total binding free energy indicates that both wild type and mutants complex formation is favorable. However, the mutants complexation is less favorable than the wild type’s. The present study provides the atomic level understanding of the aggregation behavior and the driving force for the amyloid aggregates, and could be useful for rational design of amyloid inhibitors and amyloid-specific biomarkers for diagnostic purposes.

5ns Sh-St6, M1G, 10ns Sh-St6, M1G













Similar content being viewed by others
References
Selkoe DJ (2003) Nature 426:900–904
Hamley IW (2007) Angew Chem Int Edit 46:8128–8147
Makin OS, Serpell LC (2005) Febs J 272:5950–5961
Nelson R, Eisenberg D (2006) Curr Opin Struc Biol 16:260–265
Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, Madsen AO, Riekel C, Eisenberg D (2007) Nature 447:453–457
Wiltzius JJW, Sievers SA, Sawaya MR, Cascio D, Popov D, Riekel C, Eisenberg D (2008) Protein Sci 17:1467–1474
Ivanova MI, Sievers SA, Sawaya MR, Wall JS, Eisenberg D (2009) Proc Natl Acad Sci USA 106:18990–18995
Park J, Kahng B, Hwang W (2009) PLoS Comput Biol 5:17
De Simone A, Esposito L, Pedone C, Vitagliano L (2008) Biophys J 95:1965–1973
Gsponer J, Haberthur U, Caflisch A (2003) Proc Natl Acad Sci USA 100:5154–5159
Esposito L, Paladino A, Pedone C, Vitagliano L (2008) Biophys J 94:4031–4040. doi:10.1529/biophysj.107.118935
Esposito L, Pedone C, Vitagliano L (2006) Proc Natl Acad Sci USA 103:11533–11538
Wei GH, Song W, Derreumaux P, Mousseau N (2008) Front Biosci 13:5681–5692
Wu C, Wang ZX, Lei HX, Zhang W, Duan Y (2007) J Am Chem Soc 129:1225–1232
Berryman JT, Radford SE, Harris SA (2009) Biophys J 97:1–11
Chang LK, Zhao JH, Liu HL, Liu KT, Chen JT, Tsai WB, Ho Y (2009) J Biomol Struct Dyn 26:731–740
Nerelius C, Johansson J, Sandegren A (2009) Front Biosci 14:1716–U3856
Rauk A (2008) Dalt Transact 1273-1282
Rauk A (2009) Chem Soc Rev 38:2698–2715
Teplow DB, Lazo ND, Bitan G, Bernstein S, Wyttenbach T, Bowers MT, Baumketner A, Shea JE, Urbanc B, Cruz L, Borreguero J, Stanley HE (2006) Acc Chem Res 39:635–645
Berriman J, Serpell LC, Oberg KA, Fink AL, Goedert M, Crowther RA (2003) Proc Natl Acad Sci USA 100:9034–9038
Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D (2005) Nature 435:773–778
Sunde M, Blake C (1997) Adv Prot Chem , Vol 50. Academic, San Diego, pp 123-159
Devlin GL, Knowles TPJ, Squires A, McCammon MG, Gras SL, Nilsson MR, Robinson CV, Dobson CM, MacPhee CE (2006) J Mol Biol 360:497–509
Hong DP, Fink AL (2005) Biochemistry 44:16701–16709
Wilhelm KR, Yanamandra K, Gruden MA, Zamotin V, Malisauskas M, Casaite V, Darinskas A, Forsgren L, Morozova-Roche LA (2007) Eur J Neurol 14:327–334
Brange J, Andersen L, Laursen ED, Meyn G, Rasmussen E (1997) J Pharm Sci 86:517–525
Ahmad A, Uversky VN, Hong D, Fink AL (2005) J Biol Chem 280:42669–42675
Zhang ZQ, Chen H, Bai HJ, Lai LH (2007) Biophys J 93:1484–1492
Xu WX, Ping J, Li WF, Mu YG (2009) J Chem Phys 130:8
Kayed R, Pensalfini A, Margol L, Sokolov Y, Sarsoza F, Head E, Hall J, Glabe C (2009) J Biol Chem 284:4230–4237
Quist A, Doudevski L, Lin H, Azimova R, Ng D, Frangione B, Kagan B, Ghiso J, Lal R (2005) Proc Natl Acad Sci USA 102:10427–10432
Vitagliano L, Stanzione F, De Simone A, Esposito L (2009) Biopolymers 91:1161–1171
D.A. Case TAD TEC, III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Ross C. Walker, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA (2008) University of California, San Francisco
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) J Chem Phys 103:8577–8593
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) J Comput Phys 23:327–341
Kabsch W, Sander C (1983) Biopolymers 22:2577–2637
Humphrey W, Dalke A, Schulten K (1996) J Mol Graph 14:33–38
Kollman PA, Massova I, Reyes C, Kuhn B, Huo SH, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE (2000) Acc Chem Res 33:889–897
Chong LT, Duan Y, Wang L, Massova I, Kollman PA (1999) Proc Natl Acad Sci USA 96:14330–14335
Hardy J, Selkoe DJ (2002) Science 297:353–356
Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH (2006) Nature 440:352–357
Sciarretta KL, Gordon DJ, Meredith SC (2006) Amyloid, Prions, and Other Protein Aggregates. Pt C Elsevier, San Diego, pp 273–312
Bogan AA, Thorn KS (1998) J Mol Biol 280:1–9
Haydar SN, Yun HED, Staal RGW, Hirst WD (2009) Annu Rep Med Chem, Vol 44. Elsevier, San Diego, pp 51–69
Blazer LL, Neubig RR (2009) Neuropsychopharmacology 34:126–141
Otto M, Lewczuk P, Wiltfang J (2008) Methods 44:289–298
Tenidis K, Waldner M, Bernhagen J, Fischle W, Bergmann M, Weber M, Merkle ML, Voelter W, Brunner H, Kapurniotu A (2000) J Mol Biol 295:1055–1071
Porat Y, Mazor Y, Efrat S, Gazit E (2004) Biochemistry 43:14454–14462
Sato T, Kienlen-Campard P, Ahmed M, Liu W, Li HL, Elliott JI, Aimoto S, Constantinescu SN, Octave JN, Smith SO (2006) Biochemistry 45:5503–5516
Takahashi T, Ohta K, Mihara H. Prot Struct Func Bioinf 78:336-347
Kim YS, Lee JH, Ryu J, Kim DJ (2009) Curr Pharm Des 15:637–658
Acknowledgments
This work was supported in part by the National Science Foundation (CCF/CHE 0832622). This research used resources of the National Energy Research Scientific Computing Center, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. The author appreciates the anonymous reviewers for their insightful comments, which greatly helped improving this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Berhanu, W.M., Masunov, A.E. Can molecular dynamics simulations assist in design of specific inhibitors and imaging agents of amyloid aggregation? Structure, stability and free energy predictions for amyloid oligomers of VQIVYK, MVGGVV and LYQLEN. J Mol Model 17, 2423–2442 (2011). https://doi.org/10.1007/s00894-010-0912-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-010-0912-4