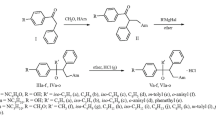
Abstract
Orthovanadate was shown to serve as a substrate for nucleoside phosphorylases from Escherichia coli, Shewanella oneidensis, Geobacillus stearothermophilus, and Halomonas chromatireducens AGD 8-3. An exception is thymidine phosphorylase from the extremophilic haloalkaliphilic bacterium Halomonas chromatireducens AGD 8-3, which cannot catalyze the vanadolysis of nucleosides. The kinetic parameters of nucleoside vanadolysis were evaluated.
Graphical abstract






Similar content being viewed by others
References
Rehder D (1992) Structure and function of vanadium compounds in living organisms. Biometals 5:3–12
Rehder D (2015) The role of vanadium in biology. Metallomics 7:730–742
Robson RL, Eady RR, Richardson TH, Miller RW, Hawkis M, Postgate JR (1986) The alternative nitrogenase of Azotobacter chroococcum is a vanadium enzyme. Nature 322:388–390
Antipov AN, Lyalikova N, Khijniak TV, L’vov NP (1998) Molybdenum-free nitrate reductases from vanadate-reducing bacteria. FEBS Lett 441:257–260
Vilter H (1984) Peroxidases from Phaeophyceae: a vanadium(V)-dependent peroxidase from Ascophyllum nodosum. Phytochemistry 23:1387–1390
Pezza RJ, Villarreal MA, Montich GG, Argarana CE (2002) Vanadate inhibits the ATPase activity and DNA binding capability of bacterial MutS. A structural model for the vanadate-MutS interaction at the Walker A motif. Nucl Acids Res 30:4700–4708
Leon-Lai CH, Gresser MJ, Tracey AS (1996) Influence of vanadium(V) complexes on the catalytic activity of ribonuclease A. The role of vanadate complexes as transition state analogues to reactions at phosphate. Can J Chem 74:38–48
Seargeant LE, Stinson RA (1979) Inhibition of human alkaline phosphatases by vanadate. Biochem J 181:247–250
McLauchlan CC, Peters BJ, Willsky GR, Crans DC (2015) Vanadium-phosphatase complexes: phosphatase inhibitors favor the trigonal bipyramidal transition state geometries. Coord Chem Rev 301–302:163–199
Davies DR, Hol WGJ (2004) The power of vanadate in crystallographic investigations of phosphoryl transfer enzymes. FEBS Lett 577:315–321
Akabayov SR, Akabayov R (2014) Vanadate in structural biology. Inorg Chim Act 420:16–23
Schramm VL (2007) Enzymatic transition state theory and transition state analogue design. J Biol Chem 282:28297–28300
Drueckhammer DG, Durrwachter JR, Pederson RL, Grans DC, Daniels L, Wong CH (1989) Reversible and in situ formation of organic arsenates and vanadates as organic phosphate mimics in enzymatic reactions: mechanistic investigation of aldol reactions and synthetic applications. J Org Chem 54:70–77
Guranowski A, Blanquet S (1986) Chromate, molybdate, tungstate and vanadate behave as substrates of yeast diadenosine 5′,5‴-p1, p4-tetraphosphate α, β-phosphorylase. Biochimie 68:757–760
Wolfe-Simon F, Blum JS, Kulp TR, Gordon GW, Hoeft SE, Pett-Ridge J, Stolz JF, Webb SM, Weber PK, Davies PC, Anbar AD, Oremland RS (2011) A bacterium that can grow by using arsenic instead of phosphorus. Science 332:1163–1166
Plass W (1999) Phosphate and vanadate in biological systems: chemical relative or more. Angew Chem 38:909–912
Evangelou AM (2002) Vanadium in cancer treatment. Crit Rev Oncol Hematol 42:249–265
Camici M, Garcia-Gil M, Pesi R, Allegrini S, Tozzi MG (2019) Purine-metabolising enzymes and apoptosis in cancer. Cancers 1354:1–27
Trevino S, Diaz A, Sanchez-Lara E, Sanchez-Gaytan BL, Perez-Aguilar JM, Gonzalez-Vergara E (2019) Vanadium in biological action: chemical, pharmacological aspects and metabolic implications in diabetes mellitus. Biol Trace Elem Res 188:68–98
Pessoa JC, Etcheverry S, Gambinoc D (2015) Vanadium compounds in medicine. Coord Chem Rev 301–302:24–48
Pessoa JC, Garribba E, Santos MFA, Santos-Silva T (2015) Vanadium and proteins: uptake, transport, structure. Coord Chem Rev 301–302:49–86
Srivastava C, Srivastava A (2004) Vanadium and the cardiovascular functions. Can J Physiol Pharmacol 82:833–839
Benitez J, Guggeri L, Tomaz I, Arrambide G, Navarro M, Pessoa JC, Garat B, Gambino D (2009) Design of vanadium mixed-ligand complexes as potential anti-protozoa agents. J Inorg Biochem 103:609–616
Neuhard J (1983) Utilization of preformed pyrimidine bases and nucleosides. In: Munch-Petersen A (ed) Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. Academic Press Inc, London, pp 95–148
Luccioni C, Beaumatin J, Bardot V, Lefrancois D (1994) Pyrimidine nucleotide metabolism in human colon carcinomas: comparison of normal tissues, primary tumors and xenografts. Int J Cancer 58:517–522
Furukawa T, Tabata S, Yamamoto M, Kawahara K, Shinsato Y, Minami K, Shimokawa M, Akiyama S (2018) Thymidine phosphorylase in cancer aggressiveness and chemoresistance. Pharm Res 132:15–20
Kanzaki A, Takebayashi Y, Bando H, Eliason JF, Watanabe S, Miyashita H, Fukumoto M, Toi M, Uchida T (2002) Expression of uridine and thymidine phosphorylase genes in human breast carcinoma. Int J Cancer 97:631–635
Matsushita S, Nitanda T, Furukawa T, Sumizawam T, Tani A, Nishimoto K, Akiba S, Miyadera K, Fukushima M, Yamada Y, Yoshida H, Kanzaki T, Akiyama S (1999) The effect of a thymidine phosphorylase inhibitor on angiogenesis and apoptosis in tumors. Cancer Res 59:1911–1916
Focher F, Spadari S (2001) Thymidine phosphorylase: a two-face Janus in anticancer chemotherapy. Curr Cancer Drug Targets 1:141–153
Bera H, Chigurupati S (2016) Recent discovery of non-nucleobase thymidine phosphorylase inhibitors targeting cancer. Eur J Med Chem 124:992–1003
Mikhailopulo IA, Miroshnikov AI (2010) New trends in nucleoside biotechnology. Acta Naturae 2:38–61
Xie X, Xia J, He K, Lu L, Xu Q, Chen N (2011) Low-molecular-mass purine nucleoside phosphorylase: characterization and application in enzymatic synthesis of nucleoside antiviral drugs. Biotech Lett 33:1107–1112
Razzel WE, Khorana HG (1958) Purification and properties of pyrimidine deoxyriboside phosphorylase from Escherichia coli. Biochem Biophys Acta 28:562–566
Kline PC, Schramm VL (1993) Purine nucleoside phosphorylase. Catalytic mechanism and transition-state analysis of the arsenolysis reaction. Biochemistry 32:13212–13219
Silva RG, Hirschi JS, Ghanem M, Murkin AS, Schramm VL (2011) Arsenate and phosphate as nucleophiles at the transition states of human purine nucleoside phosphorylase. Biochemistry 50:2701–2709
Birck MR, Schramm VL (2004) Nucleophilic participation in the transition state for human thymidine phosphorylase. J Am Chem Soc 126:2447–2453
Antipov AN, Mordkovich NN, Khijniak TV, Okorokova NA, Veiko VP (2020) Cloning of nucleoside phosphorylase genes from the extremophilic bacterium Halomonas chromatireducens AGD 8–3 with the construction of recombinant producer strains of these proteins and the study of their enzymatic properties. Appl Biochem Microbiol 56:37–43
Mordkovich NN, Antipov AN, Okorokova NA, Safonova TN, Polyakov KM, Veiko VP (2020) The nature of thermal stability of prokaryotic nucleoside phosphorylases. Appl Biochem Microbiol 56:662–670
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Kouni MH, Naguib FNM, Niedzwicki JG, Iltzsch MH, Cha S (1988) Uridine phosphorylase from Schistosoma mansoni. J Biol Chem 263:6081–6086
Shapovalova AA, Khijniak TV, Tourova TP, Muyzer G, Sorokin DY (2008) Heterotrophic denitrification at extremely high salt and pH by haloalkaliphilic Gammaproteobacteria from hypersaline soda lakes. Extremophiles 12:619–625
Shapovalova AA, Khijniak TV, Tourova TP, Sorokin DY (2009) Halomonas chromatireducens sp. nov., a new denitrifying facultatively haloalkaliphilic bacterium from soda salt marshes capable of aerobic chromate reduction. Mikrobiologiya 78:117–127
Blank JG, Hoffee PA (1975) Purification and properties of thymidine phosphorylase from Salmonella typhimurum. Arch Biochem Biophys 168:259–265
Pessoa JC (2015) Thirty years through vanadium chemistry. J Inorg Bioch 147:4–24
Benabe JE, Echegoyen LA, Pastrana B, Martinez-Maldonado M (1987) Mechanism of inhibition of glycolysis by vanadate. J Biol Chem 262:9555–9560
Caradoc-Davies TT, Cutfield SM, Lamont IL, John F, Cutfield JF (2004) Crystal structures of Escherichia coli uridine phosphorylase in two native and three complexed forms reveal basis of substrate specificity, induced conformational changes and influence of potassium. J Mol Biol 337:337–354
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Antipov, A.N., Okorokova, N.A., Safonova, T.N. et al. Vanadate as a new substrate for nucleoside phosphorylases. J Biol Inorg Chem 27, 221–227 (2022). https://doi.org/10.1007/s00775-021-01923-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-021-01923-2