Abstract
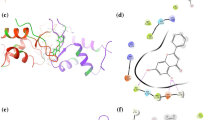
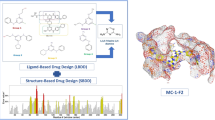
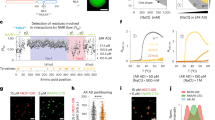
The forkhead box (FOX) transcription factor is a family of tumor suppressors that negatively regulates the tumorigenesis activity of prostate cancer; stabilization of FOX–DNA complex architecture has been recognized as a new and promising strategy for sensitizing cancer chemotherapy. Here, we described a systematic method that combined in silico analysis and in vitro assay to investigate the intermolecular interaction between FOX DNA-binding domain (DBD) and its cognate DNA partner. The structural and energetic information harvested from the molecular investigation were used to guide high-throughput virtual screening against a structurally diverse, nonredundant library of natural product compounds, aiming at discovery of novel small-molecule medicines that can conformationally stabilize and promote FOX–DNA recognition and interaction. The screening identified a number of theoretically promising hits, which were then examined by using fluorescence anisotropy assay to determine their binding potency for FOX DBD domain. The antitumor activity of identified high-affinity compounds was also tested at cellular level. Structural dynamics analysis found that the small-molecule stabilizers can shift the conformational equilibrium of FOX DBD to DNA-bound state, thus promoting the protein domain to bind tightly with its DNA partner.






Similar content being viewed by others
References
Allen WJ, Balius TE, Mukherjee S, Brozell SR, Moustakas DT, Lang PT, Case DA, Kuntz ID, Rizzo RC (2015) DOCK 6: impact of new features and current docking performance. J Comput Chem 36:1132–1156
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aoki M, Jiang H, Vogt PK (2004) Proteasomal degradation of the FoxO1 transcriptional regulator in cells transformed by the P3k and Akt oncoproteins. Proc Natl Acad Sci USA 101:13613–13617
Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O’Donovan C, Redaschi N, Yeh LS (2004) UniProt: the universal protein knowledgebase. Nucleic Acids Res 32:D115–D119
Berman H, Henrick K, Nakamura H (2003) Announcing the worldwide protein data bank. Nat Struct Biol 10:980
Butina D (1999) Unsupervised data base clustering based on Daylight’s fingerprint and Tanimoto similarity: a fast and automated way to cluster small and large data sets. J Chem Inf Model 39:747–750
Chen R, Wan J, Song J, Qian Y, Liu Y, Gu S (2017) Rational screening of peroxisome proliferator-activated receptor-γ agonists from natural products: potential therapeutics for heart failure. Pharm Biol 55:503–509
Cieplak P, Cornell WD, Bayly C, Kollman PA (1995) Application of the multimolecule and multiconformational RESP methodology to biopolymers: charge derivation for DNA, RNA, and proteins. J Comput Chem 16:1357–1377
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
DePristo MA, de Bakker PI, Blundell TL (2004) Heterogeneity and inaccuracy in protein structures solved by X-ray crystallography. Structure 12:831–838
Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA (2004) PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res 32:W665–W667
Dong C, Yan P, Wang J, Mu H, Wang S, Guo F (2017) Rational identification of natural organic compounds to target the intermolecular interaction between Foxm and DNA in colorectal cancer. Bioorg Chem 70:347–354
Good DW, Stewart GD, Hammer S, Scanlan P, Shu W, Phipps S, Reuben R, McNeill AS (2014) Elasticity as a biomarker for prostate cancer: a systematic review. BJU Int 113:523–534
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Harvey AL, Edrada-Ebel R, Quinn RJ (2015) The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov 14:111–129
Irwin JJ, Shoichet BK (2005) ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177–182
Jain RK, Mehta RJ, Nakshatri H, Idrees MT, Badve SS (2011) High-level expression of forkhead-box protein A1 in metastatic prostate cancer. Histopathology 58:766–772
Kaestner KH, Knochel W, Martinez DE (2000) Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev 14:142–146
Kamadurai HB, Subramaniam S, Jones RB, Green-Church KB, Foster MP (2003) Protein folding coupled to DNA binding in the catalytic domain of bacteriophage lambda integrase detected by mass spectrometry. Protein Sci 12:620–626
Kozakov D, Grove LE, Hall DR, Bohnuud T, Mottarella SE, Luo L, Xia B, Beglov D, Vajda S (2015) The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat Protoc 10:733–755
Lam EW, Brosens JJ, Gomes AR, Koo CY (2013) Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer 13:482–495
Larsson P, Wallner B, Lindahl E, Elofsson A (2008) Using multiple templates to improve quality of homology models in automated homology modeling. Protein Sci 17:990–1002
Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS (2003) Fox’s in development and disease. Trends Genet 19:339–344
Lu XJ, Olson WK (2003) 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res 31:5108–5121
Martin YC, Kofron JL, Traphagen LM (2002) Do structurally similar molecules have similar biological activity? J Med Chem 45:4350–4358
Myatt SS, Lam EW (2007) The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer 7:847–859
Nichols SE, Baron R, Ivetac A, McCammon JA (2011) Predictive power of molecular dynamics receptor structures in virtual screening. J Chem Inf Model 51:1439–1446
Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Wang S (2004) Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem 332:261–273
Ogrizek M, Turk S, Lešnik S, Sosič I, Hodošček M, Mirković B, Kos J, Janežič D, Gobec S, Konc J (2015) Molecular dynamics to enhance structure-based virtual screening on cathepsin B. J Comput Aided Mol Des 29:707–712
Ryckaert JP, Ciccotti G, Berendsen HJ (1997) Numerical integration of the Cartesian equations of proteins and nucleic acids. J Comput Phys 23:327–341
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Sangodkar J, Dhawan NS, Melville H, Singh VJ, Yuan E, Rana H, Izadmehr S, Farrington C, Mazhar S, Katz S, Albano T, Arnovitz P, Okrent R, Ohlmeyer M, Galsky M, Burstein D, Zhang D, Politi K, Difeo A, Narla G (2012) Targeting the FOXO1/KLF6 axis regulates EGFR signaling and treatment response. J Clin Invest 122:2637–2651
Spasic A, Serafini J, Mathews DH (2012) The Amber ff99 force field predicts relative free energy changes for RNA helix formation. J Chem Theory Comput 8:2497–2505
Sun Y, Ai X, Hou J, Ye X, Liu R, Shen S, Li Z, Lu S (2017) Integrated discovery of FOXO1-DNA stabilizers from marine natural products to restore chemosensitivity to anti-EGFR-based therapy for metastatic lung cancer. Mol BioSyst 13:330–337
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tuteja G, Kaestner KH (2007a) Forkhead transcription factors I. Cell 130:1160
Tuteja G, Kaestner KH (2007b) Forkhead transcription factors II. Cell 131:192
Tyler RC, Peterson FC, Volkman BF (2010) Distal interactions within the par3-VE-cadherin complex. Biochemistry 49:951–957
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174
Weigel D, Jäckle H (1990) The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell 63:455–456
Wright NT, Cannon BR, Wilder PT, Morgan MT, Varney KM, Zimmer DB, Weber DJ (2009) Solution structure of S100A1 bound to the CapZ peptide (TRTK12). J Mol Biol 386:1265–1277
Yang YA, Yu J (2015) Current perspectives on FOXA1 regulation of androgen receptor signaling and prostate cancer. Genes Dis 2:144–151
Yang C, Zhang S, He P, Wang C, Huang J, Zhou P (2015) Self-binding peptides: folding or binding? J Chem Inf Model 55:329–342
Yang C, Zhang S, Bai Z, Hou S, Wu D, Huang J, Zhou P (2016) A two-step binding mechanism for the selfbinding peptide recognition of target domains. Mol BioSyst 12:1201–1213
Yoshikawa S, Kukimoto-Niino M, Parker L, Handa N, Terada T, Fujimoto T, Terazawa Y, Wakiyama M, Sato M, Sano S, Kobayashi T, Tanaka T, Chen L, Liu ZJ, Wang BC, Shirouzu M, Kawa S, Semba K, Yamamoto T, Yokoyama S (2013) Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor. Oncogene 32:27–38
Zaret KS, Carroll JS (2011) Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25:2227–2241
Acknowledgements
This work was supported by The First Affiliated Hospital of Xiamen University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors confirm that this article content has no conflict of interests.
Statement of informed consent
Not applicable.
Statement of human and animal rights
Not applicable.
Additional information
Handling Editor: G. J. Peters.
Rights and permissions
About this article
Cite this article
Chen, B., Wang, H., Wu, Z. et al. Conformational stabilization of FOX–DNA complex architecture to sensitize prostate cancer chemotherapy. Amino Acids 49, 1247–1254 (2017). https://doi.org/10.1007/s00726-017-2426-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-017-2426-1