Abstract
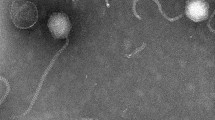
The genome sequence of a novel Meiothermus bacteriophage, named MMP7, which was isolated from Tengchong hot spring in Yunnan Province of China and belongs to the family Myoviridae, was sequenced in this study. To the best of our knowledge, this is the first reported genome sequence of a Meiothermus phage, which has a circular DNA genome of 32,864 bp and a GC content of 64%. The MMP7 genome contains 53 putative protein-encoding genes but no rRNA or tRNA genes, and it exhibits low overall sequence similarity and no significant homology to phage genomes whose sequences are publicly available, suggesting that MMP7 is a novel phage. Consistent with current taxonomic results, whole-genome-based phylogenetic analysis revealed that Meiothermus phage MMP7 has close evolutionary relationship to Thermus phages. Together, our results could be helpful for discovering new thermostable antimicrobial agents and understanding the evolution and genetic diversity of Meiothermus phages in extreme environments.


Similar content being viewed by others
References
Love MJ, Bhandari D, Dobson RCJ et al (2018) Potential for bacteriophage endolysins to supplement or replace antibiotics in food production and clinical care. Antibiotics 7:E17. https://doi.org/10.3390/antibiotics7010017
Luepke KH, Suda KJ, Boucher H et al (2017) Past, present, and future of antibacterial economics: increasing bacterial resistance, limited antibiotic pipeline, and societal implications. Pharmacotherapy 37:71–84. https://doi.org/10.1002/phar.1868
Knoll BM, Mylonakis E (2014) Antibacterial bioagents based on principles of bacteriophage biology: an overview. Clin Infect Dis 58:528–534. https://doi.org/10.1093/cid/cit771
Kutateladze M, Adamia R (2010) Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol 28:591–595. https://doi.org/10.1016/j.tibtech.2010.08.001
Gomaa S, Serry F, Abdellatif H et al (2019) Elimination of multidrug-resistant Proteus mirabilis biofilms using bacteriophages. Arch Virol 164:2265–2275. https://doi.org/10.1007/s00705-019-04305-x
Schuch R, Nelson D, Fischetti VA (2002) A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884–889. https://doi.org/10.1038/nature01026
Yang H, Yu J, Wei H (2014) Engineered bacteriophage lysins as novel anti-infectives. Front Microbiol 5:542. https://doi.org/10.3389/fmicb.2014.00542
Vazquez R, Garcia P (2019) Synergy between two chimeric lysins to kill Streptococcus pneumoniae. Front Microbiol 10:1251. https://doi.org/10.3389/fmicb.2019.01251
Wang F, Ji X, Chen M et al (2018) Rapid purification of bacteriophage endolysin TSPphg and its exogenous treatment could act as an alternative bacterial cell disruption method. Protein Expr Purif 148:54–58. https://doi.org/10.1016/j.pep.2018.03.014
Swift SM, Seal BS, Garrish JK et al (2015) A thermophilic phage endolysin fusion to a clostridium perfringens-specific cell wall binding domain creates an anti-clostridium antimicrobial with improved thermostability. Viruses 7:3019–3034. https://doi.org/10.3390/v7062758
Raposo P, Viver T, Albuquerque L et al (2019) Transfer of Meiothermus chliarophilus (Tenreiro et al. 1995) Nobre et al. 1996, Meiothermus roseus Ming et al. 2016, Meiothermus terrae Yu et al. 2014 and Meiothermus timidus Pires et al. 2005, to Calidithermus gen. nov., as Calidithermus chliarophilus comb. nov., Calidithermus roseus comb. nov., Calidithermus terrae comb. nov. and Calidithermus timidus comb. nov., respectively, and emended description of the genus Meiothermus. Int J Syst Evol Microbiol 69:1060–1069. https://doi.org/10.1099/ijsem.0.003270
Albuquerque L, Ferreira C, Tomaz D et al (2009) Meiothermus rufus sp. nov., a new slightly thermophilic red-pigmented species and emended description of the genus Meiothermus. Syst Appl Microbiol 32:306–313. https://doi.org/10.1016/j.syapm.2009.05.002
Mori K, Iino T, Ishibashi J et al (2012) Meiothermus hypogaeus sp. nov., a moderately thermophilic bacterium isolated from a hot spring. Int J Syst Evol Microbiol 62:112–117. https://doi.org/10.1099/ijs.0.028654-0
Chung AP, Rainey F, Nobre MF et al (1997) Meiothermus cerbereus sp. nov., a new slightly thermophilic species with high levels of 3-hydroxy fatty acids. Int J Syst Bacteriol 47:1225–1230. https://doi.org/10.1099/00207713-47-4-1225
Lin L, Han J, Ji X et al (2011) Isolation and characterization of a new bacteriophage MMP17 from Meiothermus. Extremophiles 15:253–258. https://doi.org/10.1007/s00792-010-0354-z
Lin L, Hong W, Ji X et al (2010) Isolation and characterization of an extremely long tail Thermus bacteriophage from Tengchong hot springs in China. J Basic Microbiol 50:452–456. https://doi.org/10.1002/jobm.201000116
Xiang X, Chen L, Huang X et al (2005) Sulfolobus tengchongensis spindle-shaped virus STSV1: virus-host interactions and genomic features. J Virol 79:8677–8686. https://doi.org/10.1128/jvi.79.14.8677-8686.2005
Bankevich A, Nurk S, Antipov D et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Aziz RK, Bartels D, Best AA et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. https://doi.org/10.1186/1471-2164-9-75
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Turner D, Reynolds D, Seto D et al (2013) CoreGenes35: a webserver for the determination of core genes from sets of viral and small bacterial genomes. BMC Res Notes 6:140. https://doi.org/10.1186/1756-0500-6-140
Santos SB, Kropinski AM, Ceyssens PJ et al (2011) Genomic and proteomic characterization of the broad-host-range Salmonella phage PVP-SE1: creation of a new phage genus. J Virol 85:11265–11273. https://doi.org/10.1128/jvi.01769-10
Acknowledgements
This study was funded by the Natural Science Foundation of China under grant no. 31760338 to FW and grant no. 31760042 to LBL, and a Start-up Grant from Kunming University of Science and Technology for the Introduction of Talent Research Project under grant no. KKSY201626003.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals requiring ethical approval.
Additional information
Handling Editor: Tim Skern.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, F., Xiao, Y., Xiong, Y. et al. Complete genome sequence of MMP7, a novel Meiothermus bacteriophage of the family Myoviridae isolated from a hot spring. Arch Virol 165, 753–756 (2020). https://doi.org/10.1007/s00705-019-04462-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04462-z