Abstract
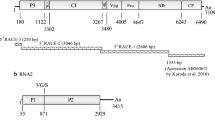
Since the first report in Costa Rica in 1971, bean rugose mosaic virus (BRMV) has been found in Colombia, El Salvador, Guatemala and Brazil. In this study, the complete genome sequence of a soybean isolate of BRMV from Paraná State, Brazil, was determined. The BRMV genome consists of two polyadenylated RNAs. RNA1 is 5909 nucleotides long and encodes a single polypeptide of 1856 amino acids (aa), with an estimated molecular weight of 210 kDa. The RNA1 polyprotein contains the polypeptides for viral replication and proteolytic processing. RNA2 is 3644 nucleotides long and codes for a single polypeptide of 1097 aa, containing the movement and coat proteins. This is the first complete genome sequence of BRMV. When compared with available aa sequences of comoviruses, the highest identities of BRMV coat proteins and proteinase polymerase were 57.5 and 58 %, respectively. These were below the 75 and 80 % identity limits, respectively, established for species demarcation in the genus. This confirms that BRMV is a member of a distinct species in the genus Comovirus.


Similar content being viewed by others
References
Almeida AMR, Ferreira LP, Henning AA, Veloso JFS, Yorinori JT (1997) Doenças Da Soja. In: Kimati H, Amorin L, Bergamin Filho A, Camargo LEA, Rezende JAM (eds) Manual de fitopatologia. Ceres, São Paulo, pp 642–664
Cupertino FP, Costa AS, Lin MT, Kitajima EW (1982) Occurrence of southern bean mosaic virus in central Brazil. Plant Dis 66:742–743
Gamez R (1982) Bean rugose mosaic virus. In: CMI/AAB descriptions of plant viruses, n. 246
Goldbach W, Wellink J (1996) Comoviruses: molecular biology and replication. In: Harrison BD, Murant AF (eds) The plant viruses. Plenum Press, New York, p 378
Khandekar S, He J, Leisner S (2009) Complete nucleotide sequence of the Toledo isolate of Turnip ringspot virus. Arch Virol 154:1917–1922
Komatsu K, Hashimoto M, Maejima K, Ozeki J, Kagiwada S, Takahashi S, Yamaji Y, Namba S (2007) Genome sequence of a Japanese isolate of Radish mosaic virus: the first complete nucleotide sequence of a crucifer-infecting Comovirus. Arch Virol 152:1501–1506
Pouwels J, Van Der Krogt GN, Van Lent J, Bisseling T, Wellink J (2002) The cytoskeleton and the secretory pathway are not involved in targeting the cowpea mosaic virus movement protein to the cell periphery. Virology 297(1):48–56
Sanfaçon H, Iwanami T, Karasev AV, Van Der Vlugt R, Wellink J, Wetzel T, Yoshikawa N (2012) Family Secoviridae. In: King AMQ, Adams MJ, Carsyens EB, Lefkowitz EJ (eds) Virus taxonomy, ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London, pp 881–899
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Wellink J, Rezelman G, Goldbach R, Beyreuther K (1986) Determination of the proteolytic processing sites in the polyprotein encoded by the bottom-competent RNA of Cowpea mosaic virus. J Virol 59:50–58
Wellink J (1998) Comovirus isolation and RNA extraction. In: Foster GD, Taylor SC (eds) Methods in molecular biology, vol 81, Plant virology protocols: from virus isolation to transgenic resistance. Humana Press, New York
Yang X, Charlebois P, Gnerre S, Coole MG, Lennon NJ, Levin JZ, Qu J, Ryan EM, Zody MC, Henn MR (2012) De novo assembly of highly diverse viral populations. BMC Genomics 13:4–5
Acknowledgments
This study was funded by Grant 472812/2012-2 of the Brazilian Research Council (CNPq).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Picoli, M.H.S., Garcia, A., Barboza, A.A.L. et al. Complete genome sequence of bean rugose mosaic virus, genus Comovirus . Arch Virol 161, 1711–1714 (2016). https://doi.org/10.1007/s00705-016-2813-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-2813-z