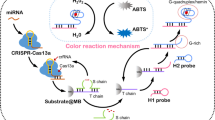
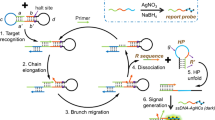
Abstract
The great selectivity and trans-cleavage activity of clustered regularly interspaced short palindromic repeats (CRISPR)/Cas13a had been coupled with high amplification efficiency of hybridization chain reaction (HCR) and magnetic-assisted enrichment, high sensitivity of electrochemiluminescence (ECL) detection to develop an ultra-sensitive biosensor for microRNA-21 (miRNA-21). The CRISPR/Cas13a was used to recognize target RNA with high specificity and performed the trans-cleavage activity. An initiation strand was generated to bind to the probe on the surface of nanomagnetic beads and then trigged HCR to produce long double-strand DNAs (dsDNAs) to realize signal amplification. Ru(phen)32+ can be inserted in the groove of the dsDNAs and acts as the ECL indicator, which can be separated through magnetic enrichment and allowed the platform to reduce the signal background. Under the optimized conditions, there is a good linear correlation between the ECL intensity and the logarithm of miRNA-21 concentration in the range 1 fM–10 nM; the limit of detection (LOD) was 0.53 fM. The proposed system was applied to detect miRNA-21 from the urine of acute kidney injury (AKI) patients with good results.
Graphical Abstract






Similar content being viewed by others
Data availability
All data in the paper are reliable. Since the majority of the experimental data were obtained from clinical samples of AKI patients, data are not publicly available according to ethical requirements.
References
Zhang W, Shu L (2016) Upregulation of miR-21 by ghrelin ameliorates ischemia/reperfusion-induced acute kidney injury by inhibiting inflammation and cell apoptosis. DNA Cell Biol 35(8):417–425. https://doi.org/10.1089/dna.2016.3231
Ledeganck KJ, Gielis EM, Abramowicz D, Stenvinkel P, Shiels PG, Van Craenenbroeck AH (2019) MicroRNAs in acute kidney injury and kidney transplantation. Clin J Am Soc Nephrol 14(3):454–468. https://doi.org/10.2215/CJN.08020718
Ishii H, Kaneko S, Yanai K, Aomatsu A, Hirai K, Ookawara S, Ishibashi K, Morishita Y (2020) MicroRNAs in podocyte injury in diabetic nephropathy. Front Genet 11:993. https://doi.org/10.3389/fgene.2020.00993
Tang J, Yao D, Yan H, Chen X, Wang L, Zhan H (2019) The role of microRNAs in the pathogenesis of diabetic nephropathy. Int J Endocrinol 8719060. https://doi.org/10.1155/2019/8719060
Li X, Pan Y, Li W, Guan P, You C (2020) The role of noncoding RNAs in gout. Endocrinology 161(11). https://doi.org/10.1210/endocr/bqaa165
Hu J, Yang Z, Wu H, Wang D (2020) Rhein attenuates renal inflammatory injury of uric acid nephropathy via lincRNA-Cox2/miR-150–5p/STAT1 axis. Int Immunopharmacol 85:106620. https://doi.org/10.1016/j.intimp.2020.106620
Magayr TA, Song X, Streets AJ, Vergoz L, Chang L, Valluru MK, Yap HL, Lannoy M, Haghighi A, Simms RJ, Tam FWK, Pei Y, Ong ACM (2020) Global microRNA profiling in human urinary exosomes reveals novel disease biomarkers and cellular pathways for autosomal dominant polycystic kidney disease. Kidney Int 98(2):420–435. https://doi.org/10.1016/j.kint.2020.02.008
Trionfini P, Benigni A, Remuzzi G (2015) MicroRNAs in kidney physiology and disease. Nat Rev Nephrol 11(1):23–33. https://doi.org/10.1038/nrneph.2014.202
Chorley BN, Ellinger-Ziegelbauer H, Tackett M, Simutis FJ, Harrill AH, McDuffie J, Atabakhsh E, Nassirpour R, Whiteley LO, Léonard JF, Carswell GK, Harpur E, Chen C, Gautier JC (2021) Urinary miRNA biomarkers of drug-induced kidney injury and their site specificity within the nephron. Toxicol Sci 180(1):1–16. https://doi.org/10.1093/toxsci/kfaa181
Pritchard CC, Cheng HH, Tewari M (2012) MicroRNA profiling: approaches and considerations. Nat Rev Genet 13(5):358–369. https://doi.org/10.1038/nrg3198
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu N, Mahuvakar VR, Andersen MR, Lao K, Livak KJ, Guegler KJ (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33(20):e179. https://doi.org/10.1093/nar/gni178
Schwarzkopf M, Pierce NA (2016) Multiplexed miRNA northern blots via hybridization chain reaction. Nucleic Acids Res 44(15):e129. https://doi.org/10.1093/nar/gkw503
Várallyay E, Burgyán J, Havelda Z (2008) MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protoc 3(2):190–196. https://doi.org/10.1038/nprot.2007.528
Jia H, Li Z, Liu C, Cheng Y (2010) Ultrasensitive detection of microRNAs by exponential isothermal amplification. Angew Chem Int Ed Engl 49(32):5498–5501. https://doi.org/10.1002/anie.201001375
Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47(D1):D155–D162. https://doi.org/10.1093/nar/gky1141
Liu H, Tian T, Zhang Y, Ding L, Yu J, Yan M (2017) Sensitive and rapid detection of microRNAs using hairpin probes-mediated exponential isothermal amplification. Biosens Bioelectron 89(Pt 2):710–714. https://doi.org/10.1016/j.bios.2016.10.099
Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary BP, Landegren U (1994) Padlock probes: circularizing oligonucleotides for localized DNA detection. Science 265(5181):2085–2088. https://doi.org/10.1126/science.7522346
Cheng Y, Zhang X, Li Z, Jiao X, Wang Y, Zhang Y (2009) Highly sensitive determination of microRNA using target-primed and branched rolling-circle amplification. Angew Chem Int Ed Engl 48(18):3268–3272. https://doi.org/10.1002/anie.200805665
Graybill RM, Bailey RC (2016) Emerging biosensing approaches for microRNA Analysis. Anal Chem 88(1):431–450. https://doi.org/10.1021/acs.analchem.5b04679
Hunt EA, Broyles D, Head T, Deo SK (2015) MicroRNA detection: current technology and research strategies. Annu Rev Anal Chem (Palo Alto Calif) 8:217–237. https://doi.org/10.1146/annurev-anchem-071114-040343
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna A, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821. https://doi.org/10.1126/science.1225829
Horvath P, Barrangou R (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327(5962):167–170. https://doi.org/10.1126/science.1179555
Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F (2016) C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353(6299):5573. https://doi.org/10.1126/science.aaf5573
Zhou T, Huang R, Huang M, Shen J, Shan Y, Xing D (2020) CRISPR/Cas13a powered portable electrochemiluminescence chip for ultrasensitive and specific miRNA detection. Adv Sci (Weinh) 7(13):1903661. https://doi.org/10.1002/advs.201903661
Liu P, Zhao K, Liu Z, Wang L, Ye S, Liang G (2021) Cas12a-based electrochemiluminescence biosensor for target amplification-free DNA detection. Biosens Bioelectron 176:112954. https://doi.org/10.1016/j.bios.2020.112954
Torrente-Rodríguez RM, Campuzano S, Montiel VR, Montoya JJ, Pingarrón JM (2016) Sensitive electrochemical determination of miRNAs based on a sandwich assay onto magnetic microcarriers and hybridization chain reaction amplification. Biosens Bioelectron 86:516–521. https://doi.org/10.1016/j.bios.2016.07.003
Xu Y, Zhang Y, Sui X, Zhang A, Liu X, Lin Z, Chen J (2021) Highly sensitive homogeneous electrochemiluminescence biosensor for microRNA-21 based on cascaded signal amplification of target-induced hybridization chain reaction and magnetic assisted enrichment. Sens. Actuators B Chem 130226. https://doi.org/10.1016/j.snb.2021.130226
Bi S, Yue S, Zhang S (2017) Hybridization chain reaction: a versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem Soc Rev 46(14):4281–4298. https://doi.org/10.1039/c7cs00055c
Lei Y, Xiao B, Liang W, Chai Y, Yuan R, Zhuo Y (2018) A robust, magnetic, and self-accelerated electrochemiluminescent nanosensor for ultrasensitive detection of copper ion. Biosens Bioelectron 109:109–115. https://doi.org/10.1016/j.bios.2018.03.013
Wang R, Zhao X, Chen X, Qiu X, Qing G, Zhang H, Zhang L, Hu X, He Z, Zhong D, Wang Y, Luo Y (2020) Rolling circular amplification (RCA)-assisted CRISPR/Cas9 cleavage (RACE) for highly specific detection of multiple extracellular vesicle MicroRNAs. Anal Chem 92(2):2176–2185. https://doi.org/10.1021/acs.analchem.9b04814
Sha Y, Huang R, Huang M, Yue H, Shan Y, Hu J, Xing D (2021) Cascade CRISPR/cas enables amplification-free microRNA sensing with fM-sensitivity and single-base-specificity. Chem Commun 57(2):247–250. https://doi.org/10.1039/d0cc06412b
Yuan C, Tian T, Sun J, Hu M, Wang X, Xiong E, Cheng M, Bao Y, Lin W, Jiang J, Yang C, Chen Q, Zhang H, Wang H, Wang X, Deng X, Liao X, Liu Y, Wang Z, Zhang G, Zhou X (2020) Universal and naked-eye gene detection platform based on the clustered regularly interspaced short palindromic Repeats/Cas12a/13a system. Anal Chem 92(5):4029–4037. https://doi.org/10.1021/acs.analchem.9b05597
Dai Y, Somoza RA, Wang L, Welter JF, Li Y, Caplan AI, Liu C (2019) Exploring the trans-cleavage activity of CRISPR-Cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew Chem Int Ed Engl 58(48):17399–17405. https://doi.org/10.1002/anie.201910772
Zhang D, Yan Y, Que H, Yang T, Cheng X, Ding S, Zhang X, Cheng W (2020) CRISPR/Cas12a-mediated interfacial cleaving of hairpin DNA reporter for electrochemical nucleic acid sensing. ACS Sens 5(2):557–562. https://doi.org/10.1021/acssensors.9b02461
Li Y, Jing Y, Hao J, Frankfort NC, Zhou X, Shen B, Liu X, Wang L, Li R (2013) MicroRNA-21 in the pathogenesis of acute kidney injury. Protein Cell 4(11):813–819. https://doi.org/10.1007/s13238-013-3085-y
Funding
This project was financially supported by the Basic and Applied Basic Research Foundation of Guangdong Province (No. 2021A1515220136) and the National Natural Science Foundation of China (No. 22174018).
Author information
Authors and Affiliations
Contributions
Yunpeng Xu: conceptualization, investigation, writing – original draft, editing. Jiahui Chen: investigation, writing – original draft. Xiaolu Sui: investigation, data curation. Yanzi Zhang: investigation. Aisha Zhang: investigation. Xinguang Liu: writing – review and editing, supervision. Zhenyu Lin: writing – review and editing, methodology, supervision. Jihong Chen: writing – review and editing, supervision.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, Y., Chen, J., Sui, X. et al. Ultra-sensitive electrochemiluminescent biosensor for miRNA based on CRISPR/Cas13a trans-cleavage-triggered hybridization chain reaction and magnetic-assisted enrichment. Microchim Acta 190, 393 (2023). https://doi.org/10.1007/s00604-023-05962-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-023-05962-1