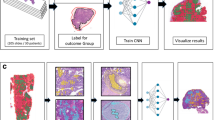
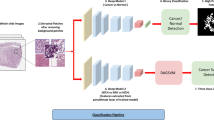
Abstract
The prognostic analysis for high grade serous adenocarcinoma (HGSC) holds significant clinical importance. However, current prognostic analysis primarily relies on statistical techniques like logistic regression and chi-square analysis alongside traditional machine learning methods based on pattern recognition. These approaches face challenges in addressing the limited reliability and validity of evaluation results, as well as the absence of reliable prognostic indicators. To identify a reliable prognostic evaluation method for high grade serous adenocarcinoma, a novel prognostic evaluation method was constructed using multi-modal deep learning techniques and compared with existing methods using data from 210 patients with high grade serous adenocarcinoma (stage III). The experimental results showed that the accuracy of this method for prognostic analysis was 80.0%, and the detection rate for poor prognosis cases was 82.87%, which was superior to current methods. Our proposed method could also automatically extract key features from different datasets and efficiently predict patient outcomes. Overall, this study laid the groundwork to overcome the difficulties in the prognostic evaluation of HGSC, help clinicians better understand the pathogenesis, and improve the long-term survival rates of this patient population.









Similar content being viewed by others
Data availability statement
All data supporting this study is available upon request by contact with the first author and corresponding authors.
References
Webb PM, Jordan SJ et al (2017) Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol 41:3–14. https://doi.org/10.1016/j.bpobgyn.2016.08.006
Morand S, Devanaboyina M, Staats H, Stanbery L, Nemunaitis J et al (2021) Ovarian cancer immunotherapy and personalized medicine. Int J Mol Sci 22(12):6532. https://doi.org/10.3390/ijms22126532
Brett MR, Jennifer BP, Thomas AS et al (2017) Epidemiology of ovarian cancer: a review. Cancer Biol Med 14(1):9–32. https://doi.org/10.20892/j.issn.2095-3941.2016.0084
Kossai ML et al (2018) Ovarian cancer: a heterogeneous disease. Pathobiology 85(1–2):41–49. https://doi.org/10.1159/000479006
Hosseini H, Monsefi R, Shadroo S et al (2022) Deep learning applications for lung cancer diagnosis: a systematic review. Electrical Eng Syst Sci 2022(01):1–32. https://doi.org/10.48550/arXiv.2201.00227
Monteiro A, Frana RP, Arthur R et al (2022) An artificial intelligent cognitive approach for classification and recognition of white blood cells employing deep learning for medical applications. Deep Learn Med Appl Unique Data 2022:53–69. https://doi.org/10.1016/b978-0-12-824145-5.00012-5
Zhang X, Wang S, Rudzinski ER et al (2022) Deep Learning of rhabdomyosarcoma pathology images for classification and survival outcome prediction. Am J Pathol Official Publ Am Assoc Pathol 192(6):917–925. https://doi.org/10.1016/j.ajpath.2022.03.011
Chen SB, Novoa RA (2022) Artificial intelligence for dermatopathology: current trends and the road ahead. Semin Diagn Pathol 39(4):298–304. https://doi.org/10.1053/j.semdp.2022.01.003
Liao X, Sun L, Yang K et al (2017) Prognostic evaluation method of ovarian granulosa cell tumor based on semi-supervised collaborative intelligence model. J Eng Sci Technol Rev 10(6):96–103. https://doi.org/10.25103/jestr.106.13
Liao X, Sun L, Yang K et al (2018) Prognosis evaluation of ovarian granulosa cell tumor based on co-forest intelligence model. J Eng Sci Technol Rev 11(2):135–142. https://doi.org/10.25103/jestr.112.19
Au KK, Josahkian JA, Francis JA, Squire JA, Koti M (2015) Current state of biomarkers in ovarian cancer prognosis. Future Oncol 11(23):3187–3195. https://doi.org/10.2217/fon.15.251
Mysona D et al (2019) A combined score of clinical factors and serum proteins can predict time to recurrence in high grade serous ovarian cancer. Gynecol Oncol 152(3):574–580. https://doi.org/10.1016/j.ygyno.2018.12.015
Clarke CL, Kushi LH, Chubak J et al (2019) Predictors of long-term survival among high-grade serous ovarian cancer patients. Cancer Epidemiol Biomarkers Prev 28(5):996–999. https://doi.org/10.1158/1055-9965.EPI-18-1324
Lisio MA, Fu L, Goyeneche A, Gao ZH, Telleria C et al (2019) High-grade serous ovarian cancer: basic sciences, clinical and therapeutic standpoints. Int J Mol Sci 20(4):952. https://doi.org/10.3390/ijms20040952
Casey L, Singh N (2019) Ovarian high-grade serous carcinoma: assessing pathology for site of origin, staging and post-neoadjuvant chemotherapy changes. Surg Pathol Clin 12(2):515–528. https://doi.org/10.1016/j.path.2019.01.007
Zeng H, Chen L, Zhang M, Luo Y, Ma X (2021) Integration of histopathological images and multi-dimensional omics analyses predicts molecular features and prognosis in high-grade serous ovarian cancer. Gynecol Oncol 163(1):171–180. https://doi.org/10.1016/j.ygyno.2021.07.015
Azzalini E, Barbazza R, Stanta G et al (2021) Histological patterns and intra-tumor heterogeneity as prognostication tools in high grade serous ovarian cancers. Gynecol Oncol 163(3):498–505. https://doi.org/10.1016/j.ygyno.2021.09.012
Yang B, Li X, Zhang W et al (2022) Spatial heterogeneity of infiltrating T cells in high-grade serous ovarian cancer revealed by multi-omics analysis. Cell Rep Med 3(12):100856. https://doi.org/10.1016/j.xcrm.2022.100856
Gayathri M, Malathy C (2022) A deep learning framework for intrusion detection and multimodal biometric image authentication. J Mobile Multimedia 18(2): 393–419. https://doi.org/10.13052/jmm1550-4646.18212
Shen K, Shi Q, Wang H et al (2021) Multimodal visibility deep learning model based on visible-infrared image pair. J Comp-Aided Des Comp Graph 33(6):939–946. https://doi.org/10.3724/SP.J.1089.2021.18420
Liu T, Huang J, Liao T et al (2021) A hybrid deep learning model for predicting molecular subtypes of human breast cancer using multimodal data. Innov Res Biomed Eng IRBM 2022(1):62–74. https://doi.org/10.1016/j.irbm.2020.12.002
Puyol-Antón E, Sidhu BS, Gould J et al (2022) A multimodal deep learning model for cardiac resynchronisation therapy response prediction. Med Image Anal 79:102465. https://doi.org/10.1016/j.media.2022.102465
Wang P, Zheng S, Jiang Y et al (2022) Structure-aware multimodal deep learning for drug-protein interaction prediction. J Chem Inf Model 62(5):1308–1317. https://doi.org/10.1021/acs.jcim.2c00060
Alattas K, Alkaabi A, Alsaud AB (2021) An overview of artificial general intelligence: recent developments and future challenges. J Comput Sci 17(4):364–370. https://doi.org/10.3844/jcssp.2021.364.370
Williams AE (2021) Approximating an artificial general intelligence or a general collective intelligence. Int J Collaborative Intell 2(3):210–223. https://doi.org/10.31730/osf.io/zsbfe
Mikki S (2023) Artificial general intelligence and noncomputability: a dynamical framework. J Artif Intell Conscious 10(01):71–101. https://doi.org/10.1142/S2705078522500163
Hygino da Cruz LC, Rodriguez I et al (2011) Pseudoprogression and pseudoresponse: imaging challen**ges in the assessment of posttreatment glioma. AJNR Am J Neuroradiol 32(11):1978–1985. https://doi.org/10.3174/ajnr.A2397
Guo Y, Zheng Z, Mao S et al (2023) Metabolic-associated signature and hub genes associated with immune microenvironment and prognosis in bladder cancer. Mol Carcinog 62(2):185–199. https://doi.org/10.1002/mc.23475
Bera K, Schalper KA, Rimm DL et al (2019) Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat Rev Clin Oncol 16(11):703–715. https://doi.org/10.1038/s41571-019-0252-y
Wan T et al (2016) A radio-genomics approach for identifying high risk estrogen receptor-positive breast cancers on DCE-MRI: preliminary results in predicting OncotypeDX risk scores. Sci Rep 6(1):21394. https://doi.org/10.1038/srep21394
McKinney SM, Sieniek M et al (2020) International evaluation of an AI system for breast cancer screening. Nature 577(7788):89–94. https://doi.org/10.1038/s41586-019-1799-6
Lao J et al (2017) A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci Rep 7(1):10353. https://doi.org/10.1038/s41598-017-10649-8
Wang L, Cao Hongrui Fu, Yang, (2022) A bearing prognosis framework based on deep wavelet extreme learning machine and particle filtering. Appl Soft Comput 131(1):109763. https://doi.org/10.1016/j.asoc.2022.109763
Bera K, Schalper KA, Rimm DL et al (2019) Artificial intelligence in digital pathology-new tools for diagnosis and precision oncology. Nat Rev Clin Onco 16:703–715. https://doi.org/10.1038/s41571-019-0252-y
Chang K, Beers AL, Bai H et al (2019). Automatic assessment of glioma burden: a deep learning algorithm for fully automated volumetric and bidimensional measurement: Neuro-oncology 21(11):1412–1422. https://doi.org/10.1093/neuonc/noz106.
da Silva Martins B, Junior RSR, Pimenta TM, de Souza JC, Rangel LBA (2022) The role of inflammasomes in ovarian cancer. In: Lele S (ed) Ovarian cancer [Internet]. Brisbane (AU): Exon Publications, 4. https://doi.org/10.36255/exon-publications-ovarian-cancer-inflammasomes
Chen H, Molberg K, Strickland AL et al (2020) PD-L1 Expression and CD8+ tumor-infiltrating lymphocytes in different types of tubo-ovarian carcinoma and their prognostic value in high-grade serous carcinoma. Am J Surg Pathol 44(8):1050–1060. https://doi.org/10.1097/PAS.0000000000001503
Koletsi D, Pandis N (2017) Survival analysis, part 2: Kaplan-Meier method and the log-rank test. Am J Orthod Dentofac Orthop 152(4):569–571. https://doi.org/10.1016/j.ajodo.2017.07.008
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. IEEE CVPR 2016:770–778. https://doi.org/10.1109/CVPR.2016.90
Tan, Mingxing and Quoc V. Le. (2019) EfficientNet: rethinking model scaling for convolutional neural networks. In: 36th International conference on machine learning: ICML 2019: 9–15. https://doi.org/10.48550/arXiv.1905.11946
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention 9351:234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Zhou Z-H, Li M (2007) Semisupervised regression with cotraining-style algorithms. IEEE Trans Knowl Data Eng 19(11):1479–1493. https://doi.org/10.1109/TKDE.2007.190644
Zhou ZH, Li M (2005) Tri-training: exploiting unlabeled data using three classifiers. IEEE Trans Knowl Data Eng 17(11):1529–1541. https://doi.org/10.1109/TKDE.2005.186
Arya N , Saha S (2021) Multi-modal advanced deep learning architectures for breast cancer survival prediction. Knowledge-Based Syst 221:106965.1–106965.11. https://doi.org/10.1016/j.knosys.2021.106965.
Yu KH, Zhang C, Berry GJ, Altman RB, Ré C et al (2016) Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 7:12474. https://doi.org/10.1038/ncomms12474
Zeng H, Chen L et al (2021) Integration of histopathological images and multi-dimensional omics analyses predicts molecular features and prognosis in high-grade serous ovarian cancer. Gynecol Oncol 163(1):171–180. https://doi.org/10.1016/j.ygyno.2021.07.015
Funding
This research was supported by National Key Research and Development Program of China (2020YFB1711500), the 1·3·5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYYC21004), the DICOM Standard National and Local Collaborated Engineering Laboratory Open Foundation (No. KFKT20220001), Provincial College Student Innovation and Entrepreneurship Training Program Project (S202310644063), and the Multi-dimensional Data Sensing and Intelligent Information Processing Key Laboratory Open Foundation (No. DWSJ2204).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Methodology, material preparation, data collection and analysis were performed by XL & KL & XZ. The first draft of the manuscript was written by XL, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no financial or proprietary interests in any material discussed in this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liao, X., Li, L., Gan, Z. et al. Prognosis prediction of high grade serous adenocarcinoma based on multi-modal convolution neural network. Neural Comput & Applic 36, 9805–9817 (2024). https://doi.org/10.1007/s00521-023-09231-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-023-09231-3