Abstract
Key message
We present a detailed protocol for isolation of single sperm cells and transcriptome analysis to study variation in gene expression between sperm cells.
Abstract
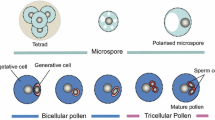
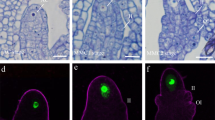
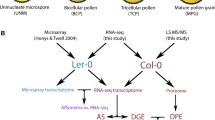
Male gametophyte development in flowering plants begins with a microspore mother cell, which upon two consecutive cell divisions forms a mature pollen grain containing a vegetative nucleus and two sperm cells. Pollen development is a highly dynamic process, involving changes at both the transcriptome and epigenome levels of vegetative nuclei and the pair of sperm cells that have their own cytoplasm and nucleus. While the overall transcriptome of Arabidopsis pollen development is well documented, studies at single-cell level, in particular of sperm cells, are still lacking. Such studies would be essential to understand whether and how the two sperm cells are transcriptionally different, in particular once the pollen tube grows through the transmitting tissue of the pistil. Here we describe a detailed protocol for isolation of single sperm cells from growing pollen tubes and analysis of their transcriptome.





Similar content being viewed by others
References
Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169
Anderson SN, Johnson CS, Jones DS, Conrad LJ, Gou X, Russell SD, Sundaresan V (2013) Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J 76:729–741
Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX, Krueger F, Smallwood SA, Ponting CP, Voet T, Kelsey G (2016) Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods 13:229–232
Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R (2015) Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10:e0128036
Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133:713–725
Becker JD, Takeda S, Borges F, Dolan L, Feijó JA (2014) Transcriptional profiling of Arabidopsis root hairs and pollen defines an apical cell growth signature. BMC Plant Biol 14:197
Berger F, Twell D (2011) Germline specification and function in plants. Ann Rev Plant Biol 62:461–484
Boavida LC, McCormick S (2007) Technical Advance: temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J 52:570–582
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD (2008) Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol 148:1168–1181
Borges F, Gardner R, Lopes T, Calarco JP, Boavida LC, Slotkin RK, Martienssen RA, Becker JD (2012) FACS-based purification of Arabidopsis microspores, sperm cells and vegetative nuclei. Plant Methods 8:44
Calarco JP, Borges F, Donoghue MTA, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijó JA, Becker JD, Martienssen RA (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151:194–205
Chen GH, Hu ZM, Guo FL, Guan XL (1995) Male gametophyte development in Plumbago zeylanica: cytoplasm localization and cell determination in 708 the early generative cell. Protoplasma 186:79–86
Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD (2017) Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89:789–804
Chumak N, Mosiolek M, Schoft VK (2015) Sample preparation and fractionation of Arabidopsis thaliana sperm and vegetative cell nuclei by FACS. Bio-protocol 5:e1664
Dupl’áková N, Dobrev PI, Reňák D, Honys D (2016) Rapid separation of Arabidopsis male gametophyte developmental stages using a Percoll gradient. Nat Protoc 11:1817
Efroni I, Birnbaum KD (2016) The potential of single-cell profiling in plants. Genome Biol 17:65
Engel ML, Chaboud A, Dumas C, McCormick S (2003) Sperm cells of Zea mays have a complex complement of mRNAs. Plant J 34:697–707
Friedman WE (1999) Expression of the cell cycle in sperm of Arabidopsis: implications for understanding patterns of gametogenesis and fertilization in plants and other eukaryotes. Development 126:1065–1075
Gawad C, Koh W, Quake SR (2016) Single-cell genome sequencing: current state of the science. Nat Rev Genet 17:175
Gou X, Yuan T, Wei X, Russell SD (2009) Gene expression in the dimorphic sperm cells of Plumbago zeylanica: transcript profiling, diversity, and relationship to cell type. Plant J 60:33–47
Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5:R85
Ibarra CA, Feng X, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, Rojas D (2012) Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337:1360–1364
Jin X, Wang RS, Zhu M, Jeon BW, Albert R, Chen S, Assmann SM (2013) Abscisic acid–responsive guard cell metabolomes of Arabidopsis wild-type and gpa1 G-protein mutants. Plant Cell 25:4789–4811
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Leydon AR, Weinreb C, Venable E, Reinders A, Ward JM, Johnson MA (2017) The molecular dialog between flowering plant reproductive partners defined by SNP-informed RNA-Sequencing. Plant Cell 29:984–1006
Libault M, Brechenmacher L, Cheng J, Xu D, Stacey G (2010) Root hair systems biology. Trends Plant Sci 15:641–650
Libault M, Pingault L, Zogli P, Schiefelbein J (2017) Plant systems biology at the single-cell level. Trends Plant Sci 22:949–960
Lin S-Y, Chen PW, Chuang MH, Juntawong P, Bailey-Serres J, Jauh GY (2014) Profiling of translatomes of in vivo-grown pollen tubes reveals genes with roles in micropylar guidance during pollination in Arabidopsis. Plant Cell 26:602–618
Liu L, Lu Y, Wei L, Yu H, Cao Y, Li Y, Yang N, Song Y, Liang C, Wang T (2018) Transcriptomics analyses reveal the molecular roadmap and long non-coding RNA landscape of sperm cell lineage development. Plant J 96:421–437
Macaulay IC, Teng MJ, Haerty W, Kumar P, Ponting CP, Voet T (2016) Separation and parallel sequencing of the genomes and transcriptomes of single cells using G&T-seq. Nat Protoc 11:2081–2103
Macaulay IC, Ponting CP, Voet T (2017) Single-cell multiomics: multiple measurements from single cells. Trends Genet 33:155–168
Mantsoki A, Devailly G, Joshi A (2016) Gene expression variability in mammalian embryonic stem cells using single cell RNA-seq data. Comput Biol Chem 63:52–61
Marinov GK, Williams BA, McCue K, Schroth GP, Gertz J, Myers RM, Wold BJ (2014) From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res 24:496–510
Marks MD, Betancur L, Gilding E, Chen F, Bauer S, Wenger JP, Dixon RA, Haigler CH (2008) A new method for isolating large quantities of Arabidopsis trichomes for transcriptome, cell wall and other types of analyses. Plant J 56:483–492
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12
Mogensen HL (1992) The male germ unit: concept, composition, and significance. Int Rev Cytol 140:129–147
Okonechnikov K, Conesa A, García-Alcalde F (2015) Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32:292–294
Palanivelu R, Preuss D (2006) Distinct short-range ovule signals attract or repel Arabidopsis thaliana pollen tubes in vitro. BMC Plant Biol 6:7
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419
Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R (2014) Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9:171
Pina C, Pinto F, Feijó JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138:744–756
Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS, Deschler DG (2017) Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171:1611–1624
Qiao Z, Libault M (2013) Unleashing the potential of the root hair cell as a single plant cell type model in root systems biology. Front Plant Sci 4:484
Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5:e1000621
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Russell SD (1985) Preferential fertilization in Plumbago: ultrastructural evidence for gamete-level recognition in an angiosperm. Proc Natl Acad Sci USA 82:6129–6132
Rutley N, Twell D (2015) A decade of pollen transcriptomics. Plant Reprod 28:73–89
Saito C, Nagata N, Sakai A, Mori K, Kuroiwa H, Kuroiwa T (2002) Angiosperm species that produce sperm cell pairs or generative cells with polarized distribution of DNA-containing organelles. Sex Plant Reprod 15:167–178
Santos MR, Bispo C, Becker JD (2017) Isolation of arabidopsis pollen, sperm cells, and vegetative nuclei by fluorescence-activated cell sorting (FACS). In: Schmidt A (ed) Plant germline development. Humana Press, New York, pp 193–210
Slotkin RK, Vaughn M, Borges F, Tanurdžić M, Becker JD, Feijó JA, Martienssen RA (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136:461–472
Twell D (2011) Male gametogenesis and germline specification in flowering plants. Sex Plant Reprod 24:149–160
Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, Jardine L (2017) Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356:eaah4573
Wang L, Wang S, Li W (2012) RSeQC: quality control of RNA-seq experiments. Bioinformatics 28:2184–2185
Yang C, Ye Z (2013) Trichomes as models for studying plant cell differentiation. Cell Mol Life Sci 70:1937–1948
Zhang J, Huang Q, Zhong S, Bleckmann A, Huang J, Guo X, Lin Q, Gu H, Dong J, Dresselhaus T, Qu LJ (2017) Sperm cells are passive cargo of the pollen tube in plant fertilization. Nat Plants 3:17079
Acknowledgements
The authors would like to acknowledge the Genomics Unit and Bioinformatics Unit of IGC. The Genomics Unit was partially supported by ONEIDA Project (LISBOA-01-0145-FEDER-016417) co-funded by FEEI—”Fundos Europeus Estruturais e de Investimento” from “Programa Operacional Regional Lisboa 2020″ and by national funds from FCT—”Fundação para a Ciência e a Tecnologia”. This work was funded by FCT ERA-CAPS project EVOREPRO (ERA-CAPS-0001-2014). CSM acknowledges a doctoral fellowship from FCT (PD/BD/114362/2016) under the Plants for Life PhD Program. JDB received salary support from FCT through an “Investigador FCT” position.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Thomas Dresselhaus.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A contribution to the special issue ‘Cellular Omics Methods in Plant Reproduction Research’.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Misra, C.S., Santos, M.R., Rafael-Fernandes, M. et al. Transcriptomics of Arabidopsis sperm cells at single-cell resolution. Plant Reprod 32, 29–38 (2019). https://doi.org/10.1007/s00497-018-00355-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-018-00355-4