Abstract
Generalizing the decomposition of a connected planar graph into a tree and a dual tree, we prove a combinatorial analog of the classic Helmholtz–Hodge decomposition of a smooth vector field. Specifically, we show that for every polyhedral complex, K, and every dimension, p, there is a partition of the set of p-cells into a maximal p-tree, a maximal p-cotree, and a collection of p-cells whose cardinality is the p-th reduced Betti number of K. Given an ordering of the p-cells, this tri-partition is unique, and it can be computed by a matrix reduction algorithm that also constructs canonical bases of cycle and boundary groups.
Similar content being viewed by others
1 Introduction
Given a connected graph embedded on the sphere, it is well known that we can split the graph into a spanning tree and a dual tree whose nodes are the faces. This is best visualized by rotating each edge that is not in the spanning tree, thus connecting the two points chosen to represent the incident faces. This split is similar in spirit to the Helmholtz decomposition of a smooth vector field on the sphere into a rotation-free component and a divergence-free component [12]. If the graph is embedded on a surface with non-zero genus, then the split does not exhaust all edges and the unused ones correspond to the third, harmonic component of the Helmholtz–Hodge decomposition on this surface [13].
Thinking of this split as a theorem about the edges of a connected planar graph, we are interested in its generalization to complexes and to cells of any dimension. Such a generalization promises a geometric interpretation of algebraic concepts in homology and their relations. Beyond this theoretical interest in the structure of complexes, we are motivated by geometric modeling tasks in which holes are of central importance. An example are cell membrane proteins with functional channels for ion transport. We believe that our structural results can be helpful in discovering and manipulating hole systems, but this is the topic of future work.
1.1 Two Examples
Our results are combinatorial and algorithmic. To get a first impression, consider the planar graph forming a wheel of \(n_0 = 17\) vertices and \(n_1 = 32\) edges drawn in the left panel of Fig. 1. Every spanning tree consists of \(n_0 - 1 = 16\) edges, and if we interpret this tree as a barrier between the \(n_2 = 17\), 2-dimensional regions defined by the embedding of the graph, then each non-tree edge splits the regions into two connected collections. In other words, the regions form a dual tree, drawn with blue edges crossing the dotted non-tree edges.
Left panel a graph of solid and dotted black edges embedded in the plane. The solid edges form a spanning tree of the graph, and the dotted edges intersect the dual blue edges, which form a spanning tree of the dual graph. Right panel an annulus decomposed into eight quadrangles. Besides a solid black spanning tree and a dotted cotree, we show the two dashed edges of a cocycle. To get a maximal cotree, we add one of the dashed edges to the cotree, while the other represents the cyclic structure of the annulus
As already noted in [1, 18], the bi-partition of the edges is best generalized to a tri-partition if the graph is embedded on a closed surface with positive genus. To further free ourselves from the implicit definition of regions, we consider complexes in which the 2-dimensional cells are explicitly specified, so it no longer matters where the complex is embedded. An example is the complex of \(n_0 = 16\) vertices, \(n_1 = 24\) edges, and \(n_2 = 8\) quadrangles drawn in the right panel of Fig. 1. An unpleasant consequence is that the dual is no longer necessarily closed as it may have edges with missing endpoints. The appropriate formalism is therefore cohomology, where we worry about edges and their incident regions rather than their incident vertices. In this formalism, a cocycle is a set of edges such that every region in the complex is incident to an even number of edges in this set, and a cotree is a set of edges that contains no non-empty cocycle. Returning to the annulus in Fig. 1, we notice that the spanning tree cannot go completely around the ‘hole’, so we get a cocycle that connects the outer boundary with the inner boundary. In other words, the complement of the spanning tree is a cotree-with-a-single-cocycle. Alternatively, we could add one edge of the cocycle to the tree to get a tree-with-a-single-cycle whose complement is a genuine cotree. Better yet, we partition the edges into a maximal tree, a maximal cotree, and one leftover edge. In the general case, there can be more leftover edges, namely one for each ‘hole’.
1.2 Results and Prior Work
The partition of the edge set has been studied for connected graphs embedded on orientable closed surfaces. Biggs proved in 1971 that such a graph splits into a spanning tree and a complementary subgraph of the dual that contains a spanning tree of the dual [1]. Rosenstiehl and Read sharpened the result in 1978 by observing that the complementary subgraph of the dual splits into a (dual) spanning tree and 2g additional edges, in which g is the genus of the surface [18]. The first result of this paper generalizes this split to complexes and to cells beyond edges. Specifically, we prove that for every polyhedral complex, K, and for every dimension, p, the set of p-cells can be partitioned into a maximal p-tree, a maximal p-cotree, and a set of leftover p-cells whose cardinality is the p-th reduced Betti number of K. An algebraic analog of this decomposition was introduced by Eckmann in 1945, namely that the p-th chain group satisfies \({{\textsf {C} }_{p}} \simeq {{\textsf {B} }_{p}} \oplus {{\textsf {B} }^{p}} \oplus {\tilde{{\textsf {H} }}_{p}}\) [6], but see also [5, 10].
The tools for establishing the tri-partition are the matrix reduction algorithms developed in the context of persistent homology [7], the theorem on the invariance of birth–death pairs proved in [3], and the duality between homology and relative cohomology noted in [19]. Importantly, the tri-partition implies canonical bases of the cycle, boundary, and homology groups as well as of their counterparts in cohomology. More specifically, for each monotonic ordering of a polyhedral complex, there is a unique tri-partition and a unique collection of bases.
1.3 Outline
Section 2 provides background from algebraic topology, including algorithms for the ranks of homology and cohomology groups. Section 3 introduces the tri-partition of a polyhedral complex. Section 4 describes the related bases in homology and cohomology and proves their properties. Section 5 concludes the paper. Appendix A proves that the tri-partitions form matroids.
2 Background
We will make frequent use of homology and cohomology groups; see [11, 16] for general background on these topics. To keep the discussion elementary, we use \({{{\mathbb {Z}}}}/ 2 {{{\mathbb {Z}}}}\) coefficients so that cycles and cocycles can be treated as sets. This choice of coefficients simplifies the arguments throughout, but it should be pointed out that some of our definitions and results depend on it. For example, the triangles of a triangulated projective plane form a 2-tree in modulo-3 arithmetic but not in modulo-2 arithmetic. For integer coefficients, there is torsion, which represents a major challenge to the generalization of our results.
2.1 Polyhedral Complexes
A p-cell, \({\sigma }\), is a p-dimensional convex polytope, and we write \({\mathrm{dim\,}}{{\sigma }} = p\) for its dimension. A hyperplane supports \({\sigma }\) if it has a non-empty intersection with the polytope and the polytope is contained in a closed half-space bounded by the hyperplane. A face of \({\sigma }\) is the intersection with a supporting hyperplane; it is a convex polytope of dimension at most p. We call \({\sigma }\) a coface of its faces. A polyhedral complex, K, is a collection of cells that is closed under taking faces such that the intersection of any two cells is a face of both. By convention, we require that the empty cell is part of K; its dimension is \(-1\) and it is a face of every cell. A cell is maximal if it has no proper coface in K. The dimension of K is the maximum dimension of any of its cells. The p-skeleton contains all cells of dimension p or less and is denoted \({{K}^{({p})}}\). We write \(K^p = {{K}^{({p})}} {\setminus } {{K}^{({p-1})}}\) for the set of p-cells in K, and \({n_{p}} = {\#\,{K^p}}\) for its cardinality, noting that \({n_{p}} = 0\) for p smaller than \(-1\) and larger than \({\mathrm{dim\,}}{K}\). The Euler characteristic is the alternating sum of cell numbers, and since the empty cell is included, we decorate it with a tilde: \({{{\tilde{\chi }}}}= \sum _p (-1)^p {n_{p}}\). The Euler–Poincaré Formula asserts that the Euler characteristic is the alternating sum of Betti numbers and therefore a topological invariant. We formally state the result now and provide the definition of the Betti numbers later.
Proposition 2.1
(Euler–Poincaré) Every polyhedral complex satisfies \({{{\tilde{\chi }}}}{=} \sum _p (-1)^p {{{\tilde{\beta }}}_{p}}\).
To represent a polyhedral complex in the computer, it is common to order the cells—arbitrarily or otherwise—and to store the face relation in a matrix form. Letting \({\sigma }_0, {\sigma }_1, \ldots , {\sigma }_m\) be the ordering, the boundary matrix, \(\partial [0 \ldots m, 0 \ldots m]\), is defined by
In words: column j of \(\partial \) stores the codimension 1 faces of \({\sigma }_j\) and row i stores the codimension 1 cofaces of \({\sigma }_i\). Throughout this paper, we use monotonic orderings in which every cell is preceded by its faces. The boundary matrix of a monotonically ordered polyhedral complex is upper-triangular. A filtration of K is a nested sequence of subcomplexes that ends with K. An example are the prefixes of a monotonic ordering: \(K_\ell = \{ {\sigma }_0, {\sigma }_1, \ldots , {\sigma }_\ell \}\) is a complex, for every \(0 \le \ell \le m\), and \(K_0 \subseteq K_1 \subseteq \cdots \subseteq K_m\) is a filtration of K.
2.2 Homology
Since we use \({{{\mathbb {Z}}}}/ 2 {{{\mathbb {Z}}}}\) coefficients, we define a p-chain as a subset of the p-cells, \({{\textsf {c} }_{p}} \subseteq K^p\). Accordingly, the sum of two p-chains is their symmetric difference, and this operation defines a group, denoted \({{\textsf {C} }_{p}} (K)\). The boundary of a p-chain is the \((p-1)\)-chain, \(\partial {{\textsf {c} }_{p}}\), that consists of all \((p-1)\)-cells shared by an odd number of p-cells in \({{\textsf {c} }_{p}}\). A p-cycle is a p-chain with empty boundary, and a p-boundary is the boundary of a \((p+1)\)-chain. Since we include the empty cell, the boundary of a vertex is this empty cell and therefore not empty. In contrast to conventional homology theory, a single vertex is therefore not a 0-cycle, but a pair of vertices is. The p-boundaries and p-cycles form subgroups of \({{\textsf {C} }_{p}}\), and because taking the boundary twice always gives the empty set, the former is a subgroup of the latter: \({{\textsf {B} }_{p}} (K) \subseteq {{\textsf {Z} }_{p}} (K) \subseteq {{\textsf {C} }_{p}} (K)\). A p-cycle in \({{\textsf {B} }_{p}} (K)\) is sometimes referred to as trivial, and two p-cycles are homologous if they differ by a p-boundary. The p-th (reduced) homology group consists of all classes of homologous p-cycles: \({\tilde{{\textsf {H} }}_{p}} (K) = {{\textsf {Z} }_{p}} (K) / {{\textsf {B} }_{p}} (K)\). The p-th (reduced) Betti number is the rank of the p-th homology group. Since we use modulo-2 arithmetic, this rank is the binary logarithm of the cardinality, and we write \({{{\tilde{\beta }}}_{p}} = {{{\tilde{\beta }}}_{p}} (K) = {\mathrm{rank\,}}{{\tilde{{\textsf {H} }}_{p}} (K)} = \log _2 {\#\,{{\tilde{{\textsf {H} }}_{p}} (K)}}\). We call K acyclic if all Betti numbers vanish. The smallest non-empty polyhedral complex is \(K = \{ \emptyset \}\), with \({{{\tilde{\beta }}}_{-1}} = 1\) and all other Betti numbers zero. The smallest non-empty acyclic polyhedral complex consists of a vertex and the empty cell.
2.3 Cohomology
While we collect information about the complex using homology, we collect information about the complement using relative cohomology; see also [19]. We begin with the definitions for cohomology. A p-cochain is a subset of the p-cells, \({{\textsf {c} }^{p}} \subseteq K^p\). Its coboundary, \(\delta {{\textsf {c} }^{p}}\), consists of all \((p+1)\)-cells that have an odd number of faces in \({{\textsf {c} }^{p}}\). A p-cocycle is a p-cochain with empty coboundary, and a p-coboundary is the coboundary of a \((p-1)\)-cochain. Again we get groups, \({{\textsf {B} }^{p}} (K) \subseteq {{\textsf {Z} }^{p}} (K) \subseteq {{\textsf {C} }^{p}} (K)\), which we distinguish from the boundary, cycle, and chain groups by writing the dimension as superscript. The p-th (reduced) cohomology group consists of all classes of cohomologous p-cocycles: \(\tilde{\textsf {H}}{}^p(K)= {{\textsf {Z} }^{p}} (K) / {{\textsf {B} }^{p}} (K)\). We write \({{{\tilde{\beta }}}^{p}} = {{{\tilde{\beta }}}^{p}} (K) = {\mathrm{rank\,}}{\tilde{\textsf {H}}{}^p(K)} = \log _2 {\#\,{\tilde{\textsf {H}}{}^p(K)}}\). For example, if \(K = \{\emptyset \}\), then \({{{\tilde{\beta }}}^{-1}} = 1\) and \({{{\tilde{\beta }}}^{p}} = 0\) for all \(p \ge 0\). We will see shortly that \({{{\tilde{\beta }}}^{p}} = {{{\tilde{\beta }}}_{p}}\) for all p, so K is acyclic iff \({{{\tilde{\beta }}}^{p}} = 0\) for all dimensions p. As a general intuition, \({{{\tilde{\beta }}}^{p}}\) is the number of cuts needed to remove all non-trivial p-th cohomology.
Relative cohomology is similar but defined for a pair, (K, L), in which L is a subcomplex of K. The relative p-cochains are the p-cochains in \(K \setminus L\), and we notice that their coboundaries are also in \(K \setminus L\). We therefore define the relative p-cocycles as the p-cocycles in \(K \setminus L\), and the relative p-coboundaries as the p-coboundaries in \(K \setminus L\). Note that this is a subtle difference to homology, where the boundary of a p-chain in \(K \setminus L\) is not necessarily in \(K \setminus L\). As before, we get three nested groups, \({{\textsf {B} }^{p}} (K,L) \subseteq {{\textsf {Z} }^{p}} (K,L) \subseteq {{\textsf {C} }^{p}} (K,L)\). The p-th (reduced) relative cohomology group is \(\tilde{\textsf {H}}{}^p(K,L) = {{\textsf {Z} }^{p}} (K,L) / {{\textsf {B} }^{p}} (K, L)\). For example, if \(L = \emptyset \), then \(\tilde{\textsf {H}}{}^p(K,L) = \tilde{\textsf {H}}{}^p(K)\), and if \(L = \{\emptyset \}\), then \(\tilde{\textsf {H}}{}^p(K,L)\) is isomorphic to the conventional cohomology group in which K does not contain the empty cell. For relative cohomology, we write \({{{\tilde{\beta }}}^{p}}(K,L) = {\mathrm{rank\,}}{\tilde{\textsf {H}}{}^p(K,L)} = \log _2 {\#\,{\tilde{\textsf {H}}{}^p(K,L)}}\).
2.4 Matrix Reduction
The classic algorithm for homology and cohomology reduces the boundary matrix to the Smith normal form; see [16, \(\S 11\)]. For modulo-2 arithmetic, this simplifies to the Gaussian elimination. We introduce special versions of this algorithm that are easy to relate to the pertinent algebraic information, including the ranks of the groups. To compute homology, we initialize \(R = \partial \) and \(U = \text{ Id }\) and reduce R using left-to-right column additions while maintaining the relation \(R = \partial U\). Write \({\mathrm{low}}{({j})}\) for the row index of the lowest non-zero item in column j of R, and set \({\mathrm{low}}{({j})} = -\infty \) if the column is zero.

We call this algorithm exhaustive because it attempts to remove non-zero entries in column j even after the lowest such entry has been established. While this strategy has also been used in [9], it is different from the standard reduction algorithm used in persistent homology, which proceeds to column \(j+1\) as soon as \({\mathrm{low}}{({j})}\) is established. An important difference is that for the exhaustive reduction algorithm, the produced matrices R and U can be uniquely defined in terms of their algebraic structure, so they do not depend on the choices of columns made by the algorithm. In contrast, the standard reduction algorithm returns matrices R and U that are generally not unique and depend on the choices of columns.
To relate the algorithm to the homology group of K, we interpret the reduction of column j as adding \({\sigma }_j\) to the complex. Since we assume a monotonic ordering, we have a polyhedral complex after every addition. There are two possible outcomes when we add \({\sigma }_j\) with \({\mathrm{dim\,}}{{\sigma }_j} = p\):
-
column j is reduced to zero, in which case \({{{\tilde{\beta }}}_{p}}\) increases by 1;
-
column j remains non-zero, in which case \({{{\tilde{\beta }}}_{p-1}}\) decreases by 1;
see [4]. In the first case, we say \({\sigma }_j\)gives birth to a p-cycle, and in the second case, we say \({\sigma }_j\)gives death to a \((p-1)\)-cycle, namely the one given birth to by \({\sigma }_i\) with \(i = {\mathrm{low}}{({j})}\); see [7, Chapter VII]. At completion, \({{{\tilde{\beta }}}_{p}}\) is the number of p-cells, \({\sigma }_j\), such that column j of R is zero and \(j \ne {\mathrm{low}}{({\ell })}\) for all \(\ell \). Writing \({n_{p}^\circ }\) for the number of p-cells that give birth and \({n_{p}^\bullet }\) for the number that give death, we have \({n_{p}} = {n_{p}^\circ } + {n_{p}^\bullet }\) and \({{{\tilde{\beta }}}_{p}} = {n_{p}^\circ } - {n_{p+1}^\bullet }\). We can therefore express \({n_{p}^\circ }\) and \({n_{p}^\bullet }\) in terms of the \({n_{q}}\) and \({{{\tilde{\beta }}}_{q}}\), and since the Betti numbers are topological invariants, we conclude that the \({n_{p}^\circ }\) and \({n_{p}^\bullet }\) neither depend on the particular reduction algorithm nor on the ordering of the cells.
To compute cohomology, we initialize \(Q = \partial \) and \(V = \text{ Id }\) and reduce Q using bottom-to-top row operations while maintaining the relation \(Q = V \partial \). Write \({\mathrm{left}}{({i})}\) for the column index of the leftmost non-zero entry in row i of Q, and set \({\mathrm{left}}{({i})} = \infty \) if the row is zero.

Similarly as before, we distinguish between two possible outcomes: that a row is reduced to zero and that it remains non-zero. To relate the algorithm to cohomology, we interpret the state of Q after reducing \({\sigma }_i\) as the relative cohomology of the pair, (K, L), in which \(L \subseteq K\) consists of the cells \({\sigma }_0\) to \({\sigma }_{i-1}\). The reduction of a row is therefore akin to moving a cell from L to \(K \setminus L\). The two possible outcomes correspond again to births and deaths, this time of relative cocycles. Writing \({n^{p}_\circ }\) and \({n^{p}_\bullet }\) for the numbers of p-cells of the two types, we have \({n_{p}} = {n^{p}_\circ } + {n^{p}_\bullet }\) and \({{{\tilde{\beta }}}^{p}} = {n^{p}_\circ } - {n^{p-1}_\bullet }\), and as before these numbers neither depend on the particular reduction algorithm nor on the ordering of the cells.
2.5 Duality
The births and deaths recorded in R and in Q are not the same but they are closely related. This is not surprising since a classic result in algebraic topology asserts that the ranks of the homology and cohomology groups coincide; see e.g. [11, 16]. We formally state this result for later reference. To relate it to the above reduction algorithms, we first prove a direct consequence of the Pairing Uniqueness Lemma in [3].
Lemma 2.2
(Pivots)Let R and Q be the matrices obtained from \(\partial \) by exhaustive column and exhaustive row reduction, respectively. Then \(i = {\mathrm{low}}{({j})}\) in R iff \(j = {\mathrm{left}}{({i})}\) in Q.
Proof
Write \(\partial _i^j = \partial [i \ldots m, 0 \ldots j]\) and define
As proved in [3], we have \(i = {\mathrm{low}}{({j})}\) in R iff \(r_\partial (i,j) = 1\). Note that this implies that \(i = {\mathrm{low}}{({j})}\) depends on \(\partial \) but not on R. Reflecting \(\partial \) across its minor diagonal, and exchanging the two indices, we get \(j = {\mathrm{left}}{({i})}\) in Q iff \(r_\partial (i, j) = 1\). The claimed equivalence follows. \(\square \)
We use Lemma 2.2 to give a short proof of the classic relation between homology and cohomology for \({{{\mathbb {Z}}}}/ 2 {{{\mathbb {Z}}}}\) coefficients.
Proposition 2.3
(Duality) Every polyhedral complex satisfies \({{{\tilde{\beta }}}_{p}} = {{{\tilde{\beta }}}^{p}}\), for all dimensions p.
Proof
We compare the ranks, which we read off the reduced boundary matrices, R and Q. Call \((\sigma _i, \sigma _j)\) a birth–death pair if \(i = {\mathrm{low}}{({j})}\) in R, which by Lemma 2.2 is equivalent to \(j = {\mathrm{left}}{({i})}\) in Q. The number of such pairs with \({\mathrm{dim\,}}{{\sigma }_i} = {\mathrm{dim\,}}{{\sigma }_j} - 1 = p-1\) is \({n_{p}^\bullet } = {n^{p-1}_\bullet }\). The p-th reduced Betti number is \({{{\tilde{\beta }}}_{p}} = {n_{p}^\circ } - {n_{p+1}^\bullet } = {n_{p}} - {n_{p}^\bullet } - {n_{p+1}^\bullet }\), and the rank of the p-th reduced cohomology group is \({{{\tilde{\beta }}}^{p}} = {n^{p}_\circ } - {n^{p-1}_\bullet } = {n_{p}} - {n^{p}_\bullet } - {n^{p-1}_\bullet }\), which we can now see are equal. \(\square \)
3 Tri-partition
This section presents the first result of this paper: the tri-partition of a polyhedral complex in which the three sets represent unique aspects of the complex’ topology. We begin with the introduction of the sets.
3.1 Trees, cotrees, and others
Letting K be a polyhedral complex, we recall that a p-chain is a subset of its p-cells. A p-tree is a p-chain, \(A_p \subseteq K^p\), that does not contain any non-empty p-cycle; compare with the definition of a generalized tree in [14]. Sometimes these generalized trees are referred to as acycles [20], which motivates our notation. A p-tree is maximal if it is not properly contained in another p-tree. Similarly, a p-cotree is a p-cochain, \(A^p \subseteq K^p\), that does not contain any non-empty p-cocycle, and it is maximal if it is not properly contained in another p-cotree. As examples consider the complexes in Fig. 1. Adding the triangles, quadrangles, and the outer face to the graph in the left panel, we get a 2-dimensional complex with 32 edges. Half the edges form a maximal 1-tree, with the other half forming a maximal 1-cotree. Moving from the left to the right panel, we get the annulus by removing the center vertex together with the incident edges and triangles as well as the outer region. The remaining 24 edges contain a maximal 1-tree of size 15 and a maximal 1-cotree of size 8, leaving one edge unused.
Our sole requirement for the third set of p-cells, \(E_p\), is that its cardinality be \({\#\,{E_p}} = {{{\tilde{\beta }}}_{p}}\). Since we talk about partitions, we have \(A_p \cap A^p = \emptyset \) and \(E_p = K^p \setminus A_p \setminus A^p\), which we will see implies the existence of \({{{\tilde{\beta }}}_{p}}\), p-cycles that generate \({\tilde{{\textsf {H} }}_{p}}\) and of \({{{\tilde{\beta }}}^{p}}\), p-cocycles that generate \(\tilde{\textsf {H}}{}^p\) such that each p-cell in \(E_p\) belongs to exactly one of these cycles and to exactly one of these cocycles; see Sect. 4 for details.
3.2 Statement and Proof
We give a constructive proof that \(K^p\) admits a tri-partition as described. More specifically, we construct such a tri-partition for every ordering of the p-cells. The ordering of the other cells is not important as long as the overall ordering of K is monotonic.
Theorem 3.1
(Tri-partition) Let K be a polyhedral complex. Then there exist tri-partitions \(A_p \sqcup A^p \sqcup E_p = K^p\), for every dimension p, such that \(A_p\) is a maximal p-tree, \(A^p\) is a maximal p-cotree, and \(E_p = K^p \setminus A_p \setminus A^p\) with \({\#\,{E_p}} = {{{\tilde{\beta }}}_{p}}\).
It is possible to argue the existence of the tri-partition in terms of column- and row-spaces of the boundary matrix. Our proof is along the same lines but more specific and designed to reveal additional properties which will be exploited in the construction of bases in Sect. 4.
Proof
We get the tri-partition in three steps: assuming a fixed monotonic ordering of K, we first construct \(A_p\), we second construct \(A^p\), and we let \(E_p\) contain the remaining p-cells.
To get started, we sort the rows and columns of the boundary matrix according to the monotonic ordering of K. For the first step, we use the exhaustive column reduction algorithm formally stated in Sect. 2. Proceeding from left to right, column j is either a combination of preceding columns, in which case it gets reduced to zero, or it is independent of the preceding columns, in which case it remains non-zero. In the latter case, we add the j-th cell to \(A_p\), in which p is the dimension of this cell. At termination, \(A_p\) is a maximal p-tree and \({\#\,{A_p}} = {n_{p}^\bullet }\) by construction.
For the second step, we use the exhaustive row reduction algorithm also formally stated in Sect. 2. Proceeding from bottom to top, row i is either a combination of succeeding (lower) rows, in which case it gets reduced to zero, or it is independent of the succeeding rows, in which case it remains non-zero. In the latter case, we add the i-th cell to \(A^p\), in which p is the dimension of this cell. At termination, \(A^p\) is a maximal p-cotree and \({\#\,{A^p}} = {n^{p}_\bullet }\) by construction.
It remains to prove that \(A_p \cap A^p = \emptyset \) and that \(E_p = K^p \setminus A_p \setminus A^p\) has cardinality \({{{\tilde{\beta }}}_{p}}\). To prove disjointness, we recall that Lemma 2.2 asserts that \(k = {\mathrm{low}}{({\ell })}\) after column reduction iff \(\ell = {\mathrm{left}}{({k})}\) after row reduction. Writing \({\sigma }_i\) for the i-th cell in the monotonic ordering, we have \({\sigma }_i \in A^p\) iff \(p = {\mathrm{dim\,}}{{\sigma }_i}\) and \({\mathrm{left}}{({i})} < \infty \). Writing \(j = {\mathrm{left}}{({i})}\), this is equivalent to \(i = {\mathrm{low}}{({j})}\), which implies that \({\sigma }_i\) gives birth to the p-cycle that \({\sigma }_j\) destroys. Hence, \({\sigma }_i \not \in A_p\), as desired. The symmetric argument shows that \(A^p\) contains no cells of \(A_p\), which implies \(A_p \cap A^p = \emptyset \). Setting \(E_p = K^p \setminus A_p \setminus A^p\), we observe that it contains a p-cell iff neither the corresponding row nor the corresponding column contains a birth–death pair. In other words, each such p-cell gives birth to an essential p-cycle in homology and, equivalently, it gives birth to an essential p-cocycle in cohomology. There are \({{{\tilde{\beta }}}_{p}} = {{{\tilde{\beta }}}^{p}}\) of each kind, hence \({\#\,{E_p}} = {{{\tilde{\beta }}}_{p}}\), as claimed. \(\square \)
The proof shows slightly more than claimed in Theorem 3.1, namely that there is a unique tri-partition for every monotonic ordering of K. On the other hand, two different monotonic orderings do not necessarily have different tri-partitions. For example, the tri-partition in dimension p is invariant as long as we retain the ordering among the p-cells, rearranging the other cells at will provided the overall ordering remains monotonic. This suggests we consider the collection of tri-partitions generated by monotonic orderings of K. Looking at its three constituents, we note that the collections of sets \(A_p\), of sets \(A^p\), and of sets \(E_p\) are three matroids; see the formal claim and the proof in Appendix A.
3.3 Tri-partitions and persistence diagrams
It is interesting to compare the tri-partition with the persistence diagram for the same monotonic ordering. Let \({\sigma }_0, {\sigma }_1, \ldots , {\sigma }_m\) be such an ordering and write \(K_\ell = \{ {\sigma }_0, {\sigma }_1, \ldots , {\sigma }_\ell \}\) for every \(0 \le \ell \le m\). The persistence diagram consists of all points (i, j) for which \(R[\,{.}\,,i] = 0\) and \(i = {\mathrm{low}}{({j})}\) and all points \((i, \infty )\) for which \(R[\,{.}\,,i] = 0\) but \(i \ne {\mathrm{low}}{({\ell })}\) for all \(0 \le \ell \le m\); see Fig. 2 and refer to [7] for details. Importantly, the number of points in upper-left quadrants anchored at points on the diagonal give the reduced Betti numbers of complexes in the filtration. Specifically, \({{{\tilde{\beta }}}_{p}} (K_\ell )\) is the number of points (i, j) with \({\mathrm{dim\,}}{{\sigma }_i} = p\) that satisfy \(i \le \ell < j\), which includes the case \(j = \infty \). As we slide the quadrant to the right and up the diagonal, we can read the reduced Betti numbers of all complexes in the filtration. With a few modifications, we can also read the ranks of the relative cohomology groups: reverse the two axes and exchange birth with death, move the points at infinity from north to west by reflecting them across the minor diagonal, and exchange the closed and open sides of the quadrant, which we slide to the left and down the diagonal.
The persistence diagram implies the tri-partition but not the other way round. Specifically, for every finite point (i, j) with \({\mathrm{dim\,}}{{\sigma }_i} = p\) in the diagram, we have \({\sigma }_i \in A^p\) and \({\sigma }_j \in A_{p+1}\), and for every point \((i, \infty )\) with \({\mathrm{dim\,}}{{\sigma }_i} = p\) we have \({\sigma }_i \in E_p\). In other words, the tri-partition records which cells give birth, which of those are essential, and which cells give death, but it does not determine the pairing that defines the persistence diagram.
3.4 Incremental Construction
We conclude this section with a brief discussion on the incremental construction of the tri-partition. Suppose we have the tri-partition of \(K_{\ell }\), how can we modify it to get the tri-partition of \(K_{\ell + 1}\)? There are only two cases, depending on whether \(R[\,{.}\,,\ell +1] = 0\) after column reduction or not. Let \(p = {\mathrm{dim\,}}{{\sigma }_{\ell +1}}\).
- Case:
-
\(R[\,{.}\,,\ell +1] = 0\). Then \({\sigma }_{\ell +1}\) gives birth to a p-cycle, so we add \({\sigma }_{\ell +1}\) to \(E_p\), leaving \(A_p\) and \(A^p\) untouched.
- Case:
-
\(R[\,{.}\,,\ell +1] \ne 0\). Then \({\sigma }_{\ell +1}\) gives death to a \((p-1)\)-cycle, and we add \({\sigma }_{\ell +1}\) to \(A_p\). Letting \(k = {\mathrm{low}}{({\ell +1})}\), we have \({\sigma }_k \in E_{p-1}\) and since its class just got killed, we move it to \(A^{p-1}\).
Note the asymmetry between the trees and the cotrees revealed by the incremental construction. Particularly perplexing, at first, is the move of \({\sigma }_k \in E_{p-1}\)—which signifies a birth in relative cohomology—to \({\sigma }_k \in A^{p-1}\)—which signifies a death in relative cohomology. The reason for this drastic change is the difference in direction, which is from left to right in the incremental construction, and from right to left in the computation of relative cohomology.
4 Bases
Besides constructing tri-partitions of a polyhedral complex, the exhaustive reduction algorithms compute canonical bases in homology and in cohomology. This section describes these bases and proves some of their properties.
4.1 Cycles and Chains
Fixing a monotonic ordering of a polyhedral complex, K, we write \(K^p = A_p \sqcup A^p \sqcup E_p\) for the corresponding tri-partition of the p-cells. For each \({\sigma }_j \in K^p\), we define a unique p-cycle or a unique p-chain with non-empty boundary. Specifically, if \({\sigma }_j \in A^p \sqcup E_p\), then there is a unique p-cycle \({{\textsf {z} }_{p}{({{\sigma }_j})}} \subseteq A_p \sqcup \{{\sigma }_j\}\), which we refer to as the canonical p-cycle of \({\sigma }_j\). If \({\sigma }_j \in A_p\), then there is a unique sum of canonical and non-trivial \((p-1)\)-cycles in \(K_{j-1}\) that is rendered trivial by adding \({\sigma }_j\). Denoting this \((p-1)\)-cycle by \({{\textsf {z} }_{}}\), there is a unique p-chain, \({{\textsf {c} }_{p}{({{\sigma }_j})}} \subseteq A_p\), with \(\partial {{\textsf {c} }_{p}{({{\sigma }_j})}} = {{\textsf {z} }_{}}\), which we refer to as the canonical p-chain of \({\sigma }_j\). Symmetrically, for every \({\sigma }_i \in K^p\), we define the canonical p-cocycle, \({{\textsf {z} }^{p}{({{\sigma }_i})}} \subseteq A^p \sqcup \{{\sigma }_i\}\), if \({\sigma }_i \in A_p \sqcup E_p\), and the canonical p-cochain, \({{\textsf {c} }^{p}{({{\sigma }_i})}} \subseteq A^p\), if \({\sigma }_i \in A^p\). We prove a technical lemma.
Lemma 4.1
(Off-diagonal Entries)Let \(R = \partial U\) and \(Q = V \partial \) be the matrix equations after exhaustive reduction. For every \(i \ne j\) there is a dimension q such that \(U[i,j] = 1\) implies \({\sigma }_i \in A_q\) and \(V[i,j] = 1\) implies \({\sigma }_j \in A^q\).
Proof
To prove that all non-zero off-diagonal entries in U belong to rows of cells in \(A_q\), for some q, we note that this is trivially true at the start of the reduction algorithm, when \(U = \text{ Id }\). Column \(\ell \) is added to column j only if \(\ell < j\) and \(R[\,{.}\,,\ell ]\) is non-zero. Since the algorithm proceeds from left to right, this implies that \({\sigma }_\ell \in A_q\). Assuming inductively that also all off-diagonal non-zero entries in \(U[\,{.}\,,\ell ]\) belong to rows of q-cells in \(A_q\), we see that the column operation maintains the claim about off-diagonal entries.
The argument why all non-zero off-diagonal entries in V belong to columns of cells in \(A^q\) is symmetric and omitted. \(\square \)
The technical lemma is useful to shed light on the connection between the canonical cycles, chains, cocycles, cochains and the matrices after exhaustive reduction.
Lemma 4.2
(Columns and Rows)After exhaustive column reduction of \(R = \partial U\), the columns of U store the canonical cycles and chains, and after exhaustive row reduction of \(Q = V \partial \), the rows of V store the canonical cocycles and cochains:
Proof
Because of symmetry, it suffices to prove the claims about the cycles and chains. Consider first the case in which \({\sigma }_j \in A^p \sqcup E_p\). After completing the reduction of column j, \(U[\,{.}\,,j]\) stores a cycle. All cells in this cycle have the same dimension as \({\sigma }_j\), which is p. Lemma 4.1 implies that this cycle is a subset of \(A_p \sqcup \{{\sigma }_j\}\). There is only one such cycle, namely \({{\textsf {z} }_{p}{({{\sigma }_j})}}\), which implies that \(U[\,{.}\,,j]\) stores this cycle, as claimed.
Consider second the case in which \({\sigma }_j \in A_p\). To show that \(U[\,{.}\,,j]\) stores \({{\textsf {c} }_{p}{({{\sigma }_j})}}\), we note that all non-zero entries in column j of U belong to rows of cells in \(A_p\), and this includes the diagonal entry. Writing \({{\textsf {c} }_{}} \subseteq A_p\) for this chain and \({{\textsf {z} }_{}} = \partial {{\textsf {c} }_{}}\) for its boundary, we note that \({{\textsf {z} }_{}}\) is stored in column j of R. Separating the birth-giving from the death-giving \((p-1)\)-cells, we write \({{\textsf {z} }_{}} = {{\textsf {z} }_{\mathrm{bth}}} \sqcup {{\textsf {z} }_{\mathrm{dth}}}\). Note that \({{\textsf {z} }_{\mathrm{bth}}} \subseteq {{\textsf {z} }_{}} \subseteq A_{p-1} \sqcup {{\textsf {z} }_{\mathrm{bth}}}\) and that there is only one such \((p-1)\)-cycle, namely the sum of the canonical \((p-1)\)-cycles of the \({\sigma }\in {{\textsf {z} }_{\mathrm{bth}}}\). Hence, \({{\textsf {z} }_{}} = \sum _{{\sigma }\in {{\textsf {z} }_{\mathrm{bth}}}} {{\textsf {z} }_{p-1}{({{\sigma }})}}\). Since the column reduction algorithm is exhaustive, each \((p-1)\)-cycle in the sum is born before \({\sigma }_j\) and dies after \({\sigma }_j\). Any other sum of non-trivial canonical \((p-1)\)-cycles is non-homologous to \({{\textsf {z} }_{}}\). By construction, \({{\textsf {z} }_{}}\) goes from non-trivial to trivial when we add \({\sigma }_j\), which implies that \(U[\,{.}\,,j]\) stores \({{\textsf {c} }_{p}{({{\sigma }_j})}}\), as claimed. \(\square \)
4.2 Canonical Bases
The columns of U and R provide bases for the cycle, boundary, and homology groups, and the rows of V and Q provide bases for the cocycle, coboundary, and cohomology groups. These bases depend on the ordering of the cells, but they are canonical in the sense that they are defined in terms of their algebraic properties and do not depend on the algorithms that compute them.
Theorem 4.3
(Canonical Bases)Assume a monotonic ordering of a polyhedral complex, K, and let \(K^p = A_p \sqcup A^p \sqcup E_p\) be the corresponding tri-partition. Then
-
\(\{ {{\textsf {z} }_{p}{({{\sigma }_j})}} \mid {\sigma }_j \in A^p \sqcup E_p \}\) is a basis of \({{\textsf {Z} }_{p}} (K)\).
-
\(\{ {{\textsf {z} }_{p}{({{\sigma }_j})}} \mid {\sigma }_j \in E_p \}\) generates a basis of \({\tilde{{\textsf {H} }}_{p}}(K)\).
-
\(\{ \partial {{\textsf {c} }_{p}{({{\sigma }_j})}} \mid {\sigma }_j \in A_p \}\) is a basis of \({{\textsf {B} }_{p-1}} (K)\).
-
\(\{ {{\textsf {z} }^{p}{({{\sigma }_i})}} \mid {\sigma }_i \in A_p \sqcup E_p \}\) is a basis of \({{\textsf {Z} }^{p}} (K)\).
-
\(\{ {{\textsf {z} }^{p}{({{\sigma }_i})}} \mid {\sigma }_i \in E_p \}\) generates a basis of
 .
. -
\(\{ \delta {{\textsf {c} }^{p}{({{\sigma }_i})}} \mid {\sigma }_i \in A^p \}\) is a basis of \({{\textsf {B} }^{p+1}} (K)\).
Proof
Because of symmetry, we can limit ourselves to the first three claims, which are about cycles and chains. We prove these claims in sequence.
To see that the \({{\textsf {z} }_{p}{({{\sigma }_j})}}\), over all \({\sigma }_j \in A^p \sqcup E_p\), form a basis of \({{\textsf {Z} }_{p}} (K)\), we note that these cycles are clearly independent. Let \({{\textsf {z} }_{}}\) be an arbitrary p-cycle, and write \({{\textsf {z} }_{j}} = {{\textsf {z} }_{p}{({{\sigma }_j})}}\) for every \({\sigma }_j \in (A^p \sqcup E_p) \cap {{\textsf {z} }_{}}\). Then \({{\textsf {z} }_{}} = \sum _j {{\textsf {z} }_{j}}\), for if they were different, then \({{\textsf {z} }_{}} + \sum _j {{\textsf {z} }_{j}}\) would be a p-cycle contained in \(A_p\), which contradicts the acyclicity of \(A_p\).
Recall that a homology class is essential if it is non-trivial in K. By construction, when \({\sigma }_j \in E_p\), then \({{\textsf {z} }_{p}{({{\sigma }_j})}}\) generates an essential class, and when \({\sigma }_j \in A^p\), then \({{\textsf {z} }_{p}{({{\sigma }_j})}}\) is trivial or homologous to a sum of cycles defined by p-cells in \(E_p\). Since \({\tilde{{\textsf {H} }}_{p}} (K)\) requires \({{{\tilde{\beta }}}_{p}}\) generators and there are only \({{{\tilde{\beta }}}_{p}}\) cells in \(E_p\), each homology class represented by such a cell must be a generator of \({\tilde{{\textsf {H} }}_{p}} (K)\).
To see that the \(\partial {{\textsf {c} }_{p}{({{\sigma }_j})}}\), over all \({\sigma }_j \in A_p\), form a basis of \({{\textsf {B} }_{p-1}} (K)\), we note that these cycles are independent. Indeed, if they were not independent, then we had a non-empty sum of chains with empty boundary, which contradicts the fact that the chains are all contained in \(A_p\) since \(A_p\) contains no cycle by construction. To show that the \(\partial {{\textsf {c} }_{p}{({{\sigma }_j})}}\) span the \((p-1)\)-dimensional boundary group, we recall that the number of \((p-1)\)-cycles \(\partial {{\textsf {c} }_{p}{({{\sigma }_j})}}\) is \({\#\,{A_p}} = {n_{p}^\bullet }\). For comparison, the rank of \({{{\textsf {B} }_{p-1}} (K)}\) is equal to the rank of \({{\textsf {Z} }_{p-1}} (K)\) minus the rank of \({\tilde{{\textsf {H} }}_{p-1}} (K)\), which is \({n_{p-1}^\circ } - ({n_{p-1}^\circ } - {n_{p}^\bullet }) = {n_{p}^\bullet }\). Since this is the same as the number of \((p-1)\)-cycles, we conclude that the \(\partial {{\textsf {c} }_{p}{({{\sigma }_j})}}\) indeed form a basis of \({{\textsf {B} }_{p-1}}\). \(\square \)
Theorem 4.3 implies the algebraic analog of the Helmholtz–Hodge decomposition, namely that the p-th chain group satisfies \({{\textsf {C} }_{p}} \simeq {{\textsf {B} }_{p-1}} \oplus {{\textsf {B} }^{p+1}} \oplus {\tilde{{\textsf {H} }}_{p}}\) for every dimension p. This is the algebraic way of saying that each p-cell either kills a \((p-1)\)-cycle, gives birth to a p-cycle that later dies, or gives birth to an essential p-cycle. Indeed, if we construct the filtration in reverse while maintaining the relative cohomology, the second of these three options corresponds to killing a relative \((p+1)\)-cocycle. To get the algebraic decomposition in a standard form, \({{\textsf {C} }_{p}} \simeq {{\textsf {B} }_{p}} \oplus {{\textsf {B} }^{p}} \oplus {\tilde{{\textsf {H} }}_{p}}\), we note \({{\textsf {B} }_{p-1}} \simeq {{\textsf {B} }^{p}}\) and \({{\textsf {B} }^{p+1}} \simeq {{\textsf {B} }_{p}}\) as needed. Indeed, the rank of \({{\textsf {B} }_{p}}\) is the number of pairs \(i = {\mathrm{low}}{({j})}\) in the column reduced boundary matrix, R, with \({\mathrm{dim\,}}{{\sigma }_i} = p\). Similarly, the rank of \({{\textsf {B} }^{p+1}}\) is the number of pairs \(j = {\mathrm{left}}{({i})}\) in the row reduced boundary matrix, Q, with \({\mathrm{dim\,}}{{\sigma }_j} = p+1\). By Lemma 2.2, the two ranks are equal and the two vector spaces are isomorphic.
4.3 Intersections of Basis Vectors
To study the relation between the various basis vectors, we consider the matrix product, VU, which we compute over \({{{\mathbb {Z}}}}\) so that \(1+1=2\).
Theorem 4.4
(Intersection Patterns)Assume a monotonic ordering of a polyhedral complex, K, and let \(K^p = A_p \sqcup A^p \sqcup E_p\) be the corresponding tri-partition in dimension p. Then
Proof
Recall that U and V are both upper-triangular, with all diagonal entries equal to 1. It follows that VU is upper-triangular, with all diagonal entries equal to 1 as well. To prove (5) for the off-diagonal entries of VU, we recall Lemma 4.1 and note that it implies
Indeed, all off-diagonal non-zero entries in row i of V belong to columns of cells in \(A^p\). Similarly, all off-diagonal non-zero entries in column j of U belong to rows of cells in \(A_p\). Multiplying the row with the column thus has the effect of adding the items at position [i, j] in the two matrices. This implies that all entries are 0, 1, or 2. We have \(V[i,j] = 1\) only if \({\sigma }_j \in A^p\), and \(U[i,j] = 1\) only if \({\sigma }_i \in A_p\). Hence, \(VU[i,j] = 2\) only if \({\sigma }_i \in A_p\) and \({\sigma }_j \in A^p\), which implies the second line of (5), and \(VU[i,j] = 1\) only if one of the two conditions is true, which implies the third line of (5). \(\square \)
Figure 3 illustrates Theorem 4.4 for \(p=1\) in a triangulated disk. The (co)cycles and (co)chains are computed with implementations of the reduction algorithms described in Sect. 2.
We draw the canonical 1-cycle of \({\sigma }_j\) in orange, the canonical 1-cocycle or 1-cochain of \({\sigma }_i\) in black, and their intersection in orange-black. From left to right: the intersection consists of two edges (\({\sigma }_j\) and \({\sigma }_i\)), of one edge (\({\sigma }_j\)), and of zero edges
Consider for example the case \(({\sigma }_i, {\sigma }_j) \in E_p \times E_p\). Then Theorem 4.4 implies \(VU[i,j] = 1\) if \(i=j\) and \(VU[i,j] = 0\) if \(i \ne j\). In words, each p-cell of \(E_p\) belongs to exactly one generating p-cycle of \({\tilde{{\textsf {H} }}_{p}}\) and to exactly one generating p-cocycle of \(\tilde{\textsf {H}}{}^p\), and there are no other intersections between the basis vectors of \({\tilde{{\textsf {H} }}_{p}}\) and the basis vectors of \(\tilde{\textsf {H}}{}^p\).
5 Discussion
The main contributions of this paper are the construction of a tri-partition of a polyhedral complex and the analysis of the corresponding bases in homology and in cohomology. For a given monotonic ordering, the tri-partition is unique and so are the corresponding bases. We mention a few questions suggested by the work reported in this paper:
-
Our constructions generalize to situations in which homology and cohomology are defined for field coefficients. Do they also generalize to non-field coefficients, for example the integers?
-
Can the analogy between the tri-partition and the Helmholtz–Hodge decomposition of a smooth vector field be used to gain insights on either side? For example, does the tri-partition lead to a fast algorithm for constructing harmonic cycles, that is, whose Laplacian is zero?
-
Can the tri-partitions be used to shed light on the stochastic properties of simplicial complexes as studied in [15]?
Applications of tri-partitions outside of mathematics are at least as important as finding connections within mathematics; see [8] for a first step. Particularly interesting is the use of the trees and cotrees to explore cave systems, such as within biomolecules and the molecular structure of materials.
References
Biggs, N.: Spanning trees of dual graphs. J. Comb. Theory Ser. B 11, 127–131 (1971)
Chen, C., Freedman, D.: Hardness results for homology localization. Discret. Comput. Geom. 45(3), 425–448 (2011)
Cohen-Steiner, D., Edelsbrunner, H., Morozov, D.: Vines and vineyards by updating persistence in linear time. In: Computational Geometry (SCG’06), pp. 119–126. ACM, New York (2006)
Delfinado, C.J.A., Edelsbrunner, H.: An incremental algorithm for Betti numbers of simplicial complexes on the \(3\)-sphere. Comput. Aided Geom. Des. 12(7), 771–784 (1995)
Dodziuk, J.: Combinatorial and continuous Hodge theories. Bull. Am. Math. Soc. 80, 1014–1016 (1974)
Eckmann, B.: Harmonische Funktionen und Randwertaufgaben in einem Komplex. Comment. Math. Helv. 17, 240–255 (1945)
Edelsbrunner, H., Harer, J.L.: Computational Topology. An Introduction. American Mathematical Society, Providence (2010)
Edelsbrunner, H., Ölsböck, K.: Holes and dependences in an ordered complex. Comput. Aided Geom. Des. 73, 1–15 (2019)
Edelsbrunner, H., Zomorodian, A.: Computing linking numbers of a filtration. Homol. Homotopy Appl. 5(2), 19–37 (2003)
Friedman, J.: Computing Betti numbers via combinatorial Laplacians. Algorithmica 21(4), 331–346 (1998)
Hatcher, A.: Algebraic Topology. Cambridge University Press, Cambridge (2002)
Helmholtz, H.: Über Integrale der hydrodynamischen Gleichungen, welche den Wirbelbewegungen entsprechen. J. Reine Angew. Math. 55, 25–55 (1858)
Hodge, W.V.D.: The Theory and Applications of Harmonic Integrals. Cambridge University Press, Cambridge (1941)
Kalai, G.: Enumeration of \(Q\)-acyclic simplicial complexes. Isr. J. Math. 45(4), 337–351 (1983)
Linial, N., Peled, Y.: Random simplicial complexes: around the phase transition. In: A Journey Through Discrete Mathematics, pp. 543–570. Springer, Cham (2017)
Munkres, J.R.: Elements of Algebraic Topology. Addison-Wesley, Menlo Park (1984)
Oxley, J.G.: Matroid Theory. Oxford Science Publications. Oxford University Press, Oxford (1992)
Rosenstiehl, P., Read, R.C.: On the principal edge tripartition of a graph. Ann. Discret. Math. 3, 195–226 (1978)
de Silva, V., Morozov, D., Vejdemo-Johansson, M.: Dualities in persistent (co)homology. Inverse Probl. 27(12), 124003 (2011)
Skraba, P., Thoppe, G., Yogeshwaran, D.: Randomly weighted \(d\)-complexes: minimal spanning acycles and persistence diagrams (2017). arXiv:1701.00239
Acknowledgements
Open access funding provided by the Institute of Science and Technology (IST Austria). The authors thank Ondřej Draganov for pointing out a mistake in an earlier version of this paper, Jörg Peters, Konrad Polthier, and Günter Rote for insightful discussions, and two anonymous reviewers for their constructive comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor in Charge: János Pach
Dedicated to the memory of Ricky Pollack.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This project has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (Grant Agreement No. 78818 Alpha). It is also partially supported by the DFG Collaborative Research Center TRR 109, ‘Discretization in Geometry and Dynamics’, through Grant No. I02979-N35 of the Austrian Science Fund (FWF).
Appendix A: Matroids
Appendix A: Matroids
The fact that the greedy algorithm succeeds in constructing maximal trees and maximal cotrees is not surprising since both form matroids [17]. We recall what this means. Let E be finite and \({{{\mathcal {F}}}}\) be a collection of subsets of E. We call \(({{{\mathcal {F}}}}, E)\) an abstract simplicial complex if \(\emptyset \in {{{\mathcal {F}}}}\) and \({{{\mathcal {F}}}}\) is closed under taking subsets. It is a matroid if, in addition, \(({{{\mathcal {F}}}}, E)\) satisfies the exchange property: \(F, G \in {{{\mathcal {F}}}}\) with \({\#\,{G}} < {\#\,{F}}\) implies the existence of \(e \in F\) such that \(G \cup \{e\} \in {{{\mathcal {F}}}}\). Traditionally, the sets in \({{{\mathcal {F}}}}\) are called independent. The exchange property implies that all maximal independent sets in \({{{\mathcal {F}}}}\) have the same cardinality. It is often convenient to focus on the maximal sets as all others are implied by inclusion. The exchange property can be replaced by the following, equivalent property: if F, G are different maximal independent sets of \({{{\mathcal {F}}}}\) and \(a \in F \setminus G\), then there exists \(b \in G \setminus F\) such that \(F \setminus \{a\} \cup \{b\} \in {{{\mathcal {F}}}}\).
Given a polyhedral complex, K, we write \({{{\mathcal {A}}}}_p\), \({{{\mathcal {A}}}}^p\), and \({{{\mathcal {E}}}}_p\) for the collections of p-trees, p-cotrees, and p-dimensional leftovers, which we recall are sets of p-cells. We note that all these sets arise in tri-partitions of K constructed for some monotonic ordering. Indeed, every \(A_p \in {{{\mathcal {A}}}}_p\) arises as a subset of the maximal p-tree if we order the p-cells in \(A_p\) before all others. Symmetrically, every \(A^p \in {{{\mathcal {A}}}}^p\) arises as a subset of the maximal p-cotree if we order the p-cells in \(A^p\) after all others. To see that every \(E_p \in {{{\mathcal {E}}}}_p\) arises as a subset of the leftover constructed for some ordering, we recall that there are collections \(D_p = \{ {{\textsf {z} }_{p}{({{\sigma }})}} \mid {\sigma }\in E_p \}\) and \(D^p = \{ {{\textsf {z} }^{p}{({{\sigma }})}} \mid {\sigma }\in E_p \}\) that satisfy Theorem 4.4; see in particular the remark following the proof of this theorem. Writing \(\bigcup D_p\) and \(\bigcup D^p\) for the p-cells that belong to the cycles and cocycles in the two collections, we note that \(\bigcup D_p \setminus E_p\) and \(\bigcup D^p \setminus E_p\) are disjoint, so we can make sure that the p-cells in \(\bigcup D_p \setminus E_p\) precede the p-cells in \(E_p\), and the latter precede the p-cells in \(\bigcup D^p \setminus E_p\). Adding the remaining p-cells arbitrarily, we get \(E_p\) as a subset of the leftover for the ordering.
It is well known that the trees and cotrees have matroid structure. We add that the same is true for the leftover sets.
Lemma A.1
(Tri-matroids) Let K be a polyhedral complex. Then \(({{{\mathcal {A}}}}_p, K^p)\), \(({{{\mathcal {A}}}}^p, K^p)\), and \(({{{\mathcal {E}}}}_p, K^p)\) are matroids for every dimension p.
Proof
We prove the claim for p-trees as a warm-up exercise, skipping the argument for p-cotrees, which is almost verbatim the same. Let \(F, G \in {{{\mathcal {A}}}}_p\) be maximal, and let \(a \in F \setminus G\). Adding a to G creates a unique p-cycle, \(A \subseteq G \cup \{a\}\). Adding a p-cell \(b \in A \setminus F\) to F creates again a unique p-cycle, which for the purpose of this proof we refer to as an elementary p-cycle in \(F \cup A\). The elementary p-cycles span the entire space of p-cycles of \(F \cup A\), which includes A. We have \(a \in A\), so a must belong to at least one elementary p-cycle. Letting \(b \in A \setminus F\) be a p-cell whose elementary p-cycle contains a, we get \(F \setminus \{a\} \cup \{b\}\) as an independent set. Noting that \(b \in G\), this implies that \(( {{{\mathcal {A}}}}_p, K^p)\) is a matroid, as claimed.
To prove that \(({{{\mathcal {E}}}}_p, K^p)\) is a matroid, we use the fact that \(({{{\mathcal {A}}}}_p, K^p)\) and \(({{{\mathcal {A}}}}^p, K^p)\) are matroids, and that for each maximal \(E_p \in {{{\mathcal {E}}}}_p\) there are maximal \(A_p \in {{{\mathcal {A}}}}_p\) and \(A^p \in {{{\mathcal {A}}}}^p\) such that \(A_p \sqcup A^p \sqcup E_p = K^p\). Let \(E_p' \in {{{\mathcal {E}}}}_p\) be maximal and different from \(E_p\), and let \(A_p'\), \({A^p}'\) be a maximal p-tree and a maximal p-cotree with \(A_p' \sqcup {A^p}' \sqcup E_p' = K^p\). Let \(a \in E_p \setminus E_p'\) and assume without loss of generality that \(a \in A_p'\). We add a to \(A_p\) and let \(b \ne a\) be any p-cell of the thus created unique p-cycle. Removing b from \(A_p \cup \{a\}\), we get again a p-tree. If \(b \in E_p'\), then we proceed to the next step, else \(b \in {A^p}'\), we add b to \(A^p\), and we iterate with a p-cell c in the thus created unique p-cocycle. Continuing this way, we eventually get a p-cell z in \(E_p'\). Indeed, every step makes \(A_p\) more similar to \(A_p'\) or it makes \(A^p\) more similar to \({A^p}'\), so the process must terminate. By construction, \(z \not \in E_p\) and \(E_p \setminus \{a\} \cup \{z\}\) is a maximal independent set of \({{{\mathcal {E}}}}_p\), which implies that \(({{{\mathcal {E}}}}_p, K^p)\) is a matroid, as claimed. \(\square \)
We note that the matroid structure implies that the greedy algorithm can be used to construct optimal trees, cotrees, and leftovers efficiently. This is in contrast to optimal bases, which for many objective functions are NP-hard to construct [2].
The proof that \(({{{\mathcal {E}}}}_p, K^p)\) is a matroid extends to general partitions of a ground-set. Fixing an integer k and a set E, we consider partitions \(E = F_1 \sqcup F_2 \sqcup \ldots \sqcup F_k\) such that for each \(1 \le i < k\) the collection of sets \(F_i\) is a matroid over E, and conclude that the collection of sets \(F_k\) is also a matroid over E.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Edelsbrunner, H., Ölsböck, K. Tri-partitions and Bases of an Ordered Complex. Discrete Comput Geom 64, 759–775 (2020). https://doi.org/10.1007/s00454-020-00188-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00454-020-00188-x



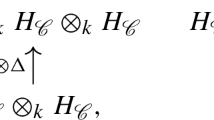


 .
.