Abstract
Copy number variations (CNVs) have provided a dynamic aspect to the apparently static human genome. We have analyzed CNVs larger than 100 kb in 477 healthy individuals from 26 diverse Indian populations of different linguistic, ethnic and geographic backgrounds. These CNVRs were identified using the Affymetrix 50K Xba 240 Array. We observed 1,425 and 1,337 CNVRs in the deletion and amplification sets, respectively, after pooling data from all the populations. More than 50% of the genes encompassed entirely in CNVs had both deletions and amplifications. There was wide variability across populations not only with respect to CNV extent (ranging from 0.04–1.14% of genome under deletion and 0.11–0.86% under amplification) but also in terms of functional enrichments of processes like keratinization, serine proteases and their inhibitors, cadherins, homeobox, olfactory receptors etc. These did not correlate with linguistic, ethnic, geographic backgrounds and size of populations. Certain processes were near exclusive to deletion (serine proteases, keratinization, olfactory receptors, GPCRs) or duplication (homeobox, serine protease inhibitors, embryonic limb morphogenesis) datasets. Populations having same enriched processes were observed to contain genes from different genomic loci. Comparison of polymorphic CNVRs (5% or more) with those cataloged in Database of Genomic Variants revealed that 78% (2473) of the genes in CNVRs in Indian populations are novel. Validation of CNVs using Sequenom MassARRAY revealed extensive heterogeneity in CNV boundaries. Exploration of CNV profiles in such diverse populations would provide a widely valuable resource for understanding diversity in phenotypes and disease.






Similar content being viewed by others
References
Abdulla MA, Ahmed I, Assawamakin A et al (2009) Mapping human genetic diversity in Asia. Science 326:1541–1545
Basu A, Mukherjee N, Roy S, Sengupta S, Banerjee S, Chakraborty M, Dey B, Roy M, Roy B, Bhattachatyya NP et al (2003) Ethinic India:a genomic view, with special reference to peopling and structure. Genome Res 13:2277–2290
Caceres A et al (2010) Multiple correspondence discriminant analysis: an application to detect stratification in copy number variation. Stat. Med. 29:3284–3293
Cann RL (2001) Genetic clues to dispersal in human populations: retracing the past from the present. Science 291:1742–1748
Chao J, Shen B, Gao L, Xia CF, Bledsoe G, Chao L (2010) Tissue kallikrein in cardiovascular, cerebrovascular and renal diseases and skin wound healing. Biol Chem 391:345–355
Clevert, Djork-Arné, Mitterecker A, Mayr, et al. (2010) cn.FARMS: a probabilistic model to detect DNA copy numbers. Nucleic Acids Research 2011:1–13
Conrad DF, Pinto D, Redon R, Feuk L et al (2010) Origins and functional impact of copy number variation in the human genome. Nature 464:704–712
Craddock N, Hurles ME, Cardin N et al (2010) Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464:713–720
Ding C, Cantor CR (2003) A high-throughput gene expression analysis technique using competitive PCR and matrix-assisted laser desorption ionization time-of-flight MS. Proc Natl Acad Sci U S A 100:3059–3064
Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH (2010) Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 11:446–450
Estivill X, Armengol L (2007) Copy number variants and common disorders: filling the gaps and exploring complexity in genome-wide association studies. PLoS Genet 3:1787–1799
Fanciulli M, Norsworthy PJ, Petretto E et al (2007) FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet 39:721–723
Frazer KA, Murray SS, Schork NJ, Topol EJ (2009) Human genetic variation and its contribution to complex traits. Nat Rev Genet 10:241–251
Gonzalez E, Kulkarni H, Bolivar H et al (2005) The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307:1434–1440
Hasin Y, Olender T, Khen M, Gonzaga-Jauregui C, Kim PM, Urban AE, Snyder M, Gerstein MB, Lancet D, Korbel JO (2008) High-resolution copy-number variation map reflects human olfactory receptor diversity and evolution. PLoS Genet 4:e1000249
Hasin-Brumshtein Y, Lancet D, Olender T (2009) Human olfaction: from genomic variation to phenotypic diversity. Trends Genet 25:178–184
Heutinck KM, ten Berge IJ, Hack CE, Hamann J, Ro wshani AT (2010) Serine proteases of the human immune system in health and disease. Mol Immunol 47(11–12):1943–1955
Huang dW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C (2004) Detection of large-scale variation in the human genome. Nat Genet 36:949–951
Indian Consortium Genome Variation (2008) Genetic landscape of the people of India: a canvas for disease gene exploration. J Genet 87:3–20
Indian Genome Variation Consortium (2005) The Indian Genome Variation database (IGVdb): a project overview. Hum Genet 118:1–11
Itsara A, Cooper GM, Baker C et al (2009) Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet 84:148–161
Jakobsson M, Scholz SW, Scheet P et al (2008) Genotype, haplotype and copy-number variation in worldwide human populations. Nature 451:998–1003
Kim PM, Korbel JO, Gerstein MB (2007) Positive selection at the protein network periphery: evaluation in terms of structural constraints and cellular context. Proc Natl Acad Sci USA 104:20274–20279
Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, Craddock N, Owen MJ, O’Donovan MC (2009) Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet 18:1497–1503
Kusenda M, Sebat J (2008) The role of rare structural variants in the genetics of autism spectrum disorders. Cytogenet Genome Res 123:36–43
Lee JA, Carvalho CM, Lupski JR (2007) A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131:1235–1247
Lopez CC, Brems H, Lazaro C, Estivill X, Clementi M, Mason S, Rutkowski JL, Marynen P, Legius E (1999) Molecular studies in 20 submicroscopic neurofibromatosis type 1 gene deletions. Hum Mutat 14:387–393
Majumder PP (1998) people of India: biological diversity and affinities. Evol Anthrop 6:100–110
Majumder PP (2001) Ethnic populations of India as seen from an evolutionary perspective. J Biosci 26:533–545
Malhotra KC (1978) Morphological composition of the people of India. J Hum Evol 7:45–63
McCarroll SA, Altshuler DM (2007) Copy-number variation and association studies of human disease. Nat Genet 39:S37–S42
McCarroll SA, Kuruvilla FG, Korn JM et al (2008) Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet 40:1166–1174
McKinney C, Fanciulli M, Merriman ME et al. (2010) Association of variation in Fc {gamma} receptor 3B gene copy number with rheumatoid arthritis in Caucasian samples. Ann Rheum Dis
Perry GH, Dominy NJ, Claw KG et al (2007) Diet and the evolution of human amylase gene copy number variation. Nat Genet 39:1256–1260
Redon R, Ishikawa S, Fitch KR, Feuk L et al (2006) Global variation in copy number in the human genome. Nature 444:444–454
Reich D, Thangaraj K, Patterson N, Price AL, Singh L (2009) Reconstructing Indian population history. Nature 461:489–494
Sebat J, Lakshmi B, Troge J, Alexander J et al (2004) Large-scale copy number polymorphism in the human genome. Science 305:525–528
Sebat J, Lakshmi B, Malhotra D et al (2007) Strong association of de novo copy number mutations with autism. Science 316:445–449
Sengupta S, Zhivotovsky LA, King R, Mehdi SQ, Edmonds CA, Chow CE, Lin AA, Mitra M, Sil SK, Ramesh A et al (2006) Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet 78:202–221
Singh KS (2002) People of India: introduction national series. Anthropological Survey of India. Oxford University Press, Delhi
The HUGO Pan-Asian SNP Consortium (2009) Mapping human genetic diversity in Asia. Science, pp 1541–1545
Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, Olson MV, Eichler EE (2005) Fine-scale structural variation of the human genome. Nat Genet 37:727–732
Walsh T, McClellan JM, McCarthy SE et al (2008) Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320:539–543
Acknowledgments
We thank Amit Chaurasia for computational and Ankita and Rishi Das Roy for IGV browser support; Financial support to MM CSIR(CMM0016, SIP0006) and Council for Scientific and Industrial Research SRF to PG and PJ is acknowledged. We also acknowledge The Centre for Genomic Applications for Microarray and Sequenom facility and Spinco Biotech Pvt. Ltd. for support with the SVS7 software. The data is available at http://igvbrowser.igib.res.in.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
P. Gautam and P. Jha contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
439_2011_1050_MOESM1_ESM.tif
Size distribution of CNVRs in Database of Genome Variants (DGV).The size distribution of segments in DGV shows that 23% of the segments are of size equal to or more than 100 kb implying the importance and prevalence of large CNVs in genome Supplementary material 1 (TIFF 1.27 Mb)
439_2011_1050_MOESM2_ESM.tif
Geographic locations of populations sampled. The populations sampled in this study cover the length and breadth of India and are from various ethnic, linguistic backgrounds Supplementary material 2 (TIFF 2817 kb)
439_2011_1050_MOESM3_ESM.tif
Inter-probe distance of Affymetrix 50K Xba array. This plot shows that around 35% of probe-pairs are 5kb apart from each other, and 25% of probe pairs are just 1kb apart giving confidence over calling CN altered regions Supplementary material 3 (TIFF 1203 kb)
439_2011_1050_MOESM4_ESM.docx
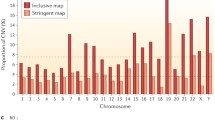
Chromosomal CNV landscape in all the populations. The 26 different population show different extent of CNVs. The red line depicts deletion and blue line amplification Supplementary material 4 (DOCX 1.91 mb)
439_2011_1050_MOESM5_ESM.tif
Multiple correspondence discriminant analysis (MCDA): multiple correspondence discriminant analysis (MCDA) on all 26 Indian populations using 632 polymorphic CNVRs (present in more than 10% of cohort) to detect population stratification Supplementary material 5 (TIFF 3590 kb)
439_2011_1050_MOESM6_ESM.docx
Heterogeneity in CNV boundary: Representation of deletion and amplification regions encompassing genes in different samples as revealed by array data. The target for Sequenom probe is indicated by black arrow. There is an enormous heterogeneity in CNV boundaries and some of the CNV regions are not queried by the Sequenom MassARRAY probe Supplementary material 6 (DOCX 2.27 mb)
439_2011_1050_MOESM7_ESM.doc
Heterogeneity in CNV boundaries in DGV: Heterogeneity in CNV boundaries as present in public database DGV for some of the genes (ABCC1, ODAM, PRKG1 and SDK1) from our validation genes set. This observation also pointed out in our data indicates the difficulties posed in the validation of such loci Supplementary material 7 (DOC 407 kb)
Rights and permissions
About this article
Cite this article
Gautam, P., Jha, P., Kumar, D. et al. Spectrum of large copy number variations in 26 diverse Indian populations: potential involvement in phenotypic diversity. Hum Genet 131, 131–143 (2012). https://doi.org/10.1007/s00439-011-1050-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-011-1050-5