Abstract
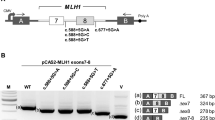
Single base substitutions in DNA mismatch repair genes which are predicted to lead either to missense or silent mutations, or to intronic variants outside the highly conserved splicing region are often found in hereditary nonpolyposis colorectal cancer (HNPCC) families. In order to use the variants for predictive testing in persons at risk, their pathogenicity has to be evaluated. There is growing evidence that some substitutions have a detrimental influence on splicing. We examined 19 unclassified variants (UVs) detected in MSH2 or MLH1 genes in patients suspected of HNPCC for expression at RNA level. We demonstrate that 10 of the 19 UVs analyzed affect splicing. For example, the substitution MLH1,c.2103G>C in the last position of exon 18 does not result in a missense mutation as theoretically predicted (p.Gln701His), but leads to a complete loss of exon 18. The substitution MLH1,c.1038G>C (predicted effect p.Gln346His) leads to complete inactivation of the mutant allele by skipping of exons 10 and 11, and by activation of a cryptic intronic splice site. Similarly, the intronic variant MLH1,c.306+2dupT results in loss of exon 3 and a frameshift mutation due to a new splice donor site 5 bp upstream. Furthermore, we confirmed complete exon skipping for the mutations MLH1,c.1731G>A and MLH1,c.677G>A. Partial exon skipping was demonstrated for the mutations MSH2,c.1275A>G, MLH1,c.588+5G>A, MLH1,c.790+4A>G and MLH1,c.1984A>C. In contrast, five missense mutations (MSH2,c.4G>A, MSH2,c.2123T>A, MLH1,c.464T>G, MLH1,c.875T>C and MLH1,c.2210A>T) were found in similar proportions in the mRNA as in the genomic DNA. We conclude that the mRNA examination should precede functional tests at protein level.



Similar content being viewed by others
References
Aretz S, Uhlhaas S, Sun Y, Pagenstecher C, Mangold E, Caspari R, Moslein G, Schulmann K, Propping P, Friedl W (2004) Familial adenomatous polyposis: aberrant splicing due to missense or silent mutations in the APC gene. Hum Mutat 24:370–380
Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3:285–298
Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 31:3568–3571
Ellison AR, Lofing J, Bitter GA (2001) Functional analysis of human MLH1 and MSH2 missense variants and hybrid human–yeast MLH1 proteins in Saccharomyces cerevisiae. Hum Mol Genet 10:1889–1900
Gorlov IP, Gorlova OY, Frazier ML, Amos CI (2003) Missense mutations in hMLH1 and hMSH2 are associated with exonic splicing enhancers. Am J Hum Genet 73(5):1157–1161
Guerrette S, Acharya S, Fishel R (1999) The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J Biol Chem 274(10):6336–6341
Kohonen-Corish M, Ross VL, Doe WF, Kool DA, Edkins E, Faragher I, Wijnen J, Khan PM, Macrae F, St John DJ (1996) RNA-based mutation screening in hereditary nonpolyposis colorectal cancer. Am J Hum Genet 59:818–824
Kondo E, Suzuki H, Horii A, Fukushige S (2003) A yeast two-hybrid assay provides a simple way to evaluate the vast majority of hMLH1 germ-line mutations. Cancer Res 63:3302–3308
Kruger S, Plaschke J, Pistorius S, Jeske B, Haas S, Kramer H, Hinterseher I, Bier A, Kreuz FR, Theissig F, Saeger HD, Schackert HK (2002) Seven novel MLH1 and MSH2 germline mutations in hereditary nonpolyposis colorectal cancer. Hum Mutat 19:82
Lastella P, Resta N, Miccolis I, Quagliarella A, Guanti G, Stella A (2004) Site directed mutagenesis of hMLH1 exonic splicing enhancers does not correlate with splicing disruption. J Med Genet 41:e72
Lynch HT, de la Chapelle A (1999) Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet 36:801–818
Mangold E, Pagenstecher C, Friedl W, Mathiak M, Buettner R, Engel C, Loeffler M, Holinski-Feder E, Muller-Koch Y, Keller G, Schackert HK, Kruger S, Goecke T, Moeslein G, Kloor M, Gebert J, Kunstmann E, Schulmann K, Ruschoff J, Propping P (2005) Spectrum and frequencies of mutations in MSH2 and MLH1 identified in 1,721 German families suspected of hereditary nonpolyposis colorectal cancer. Int J Cancer 116:692–702
Nystrom-Lahti M, Perrera C, Raschle M, Panyushkina-Seiler E, Marra G, Curci A, Quaresima B, Costanzo F, D’Urso M, Venuta S, Jiricny J (2002) Functional analysis of MLH1 mutations linked to hereditary nonpolyposis colon cancer. Genes Chromosomes Cancer 33(2):160–167
Peltomaki P (2005) Lynch syndrome genes. Fam Cancer 4(3):227–232
Raevaara TE, Korhonen MK, Lohi H, Hampel H, Lynch E, Lonnqvist KE, Holinski-Feder E, Sutter C, McKinnon W, Duraisamy S, Gerdes AM, Peltomaki P, Kohonen-Ccorish M, Mangold E, Macrae F, Greenblatt M, de la Chapelle A, Nystrom M (2005) Functional significance and clinical phenotype of nontruncating mismatch repair variants of MLH1. Gastroenterology 129(2):537–549
Sharp A, Pichert G, Lucassen A, Eccles D (2004) RNA analysis reveals splicing mutations and loss of expression defects in MLH1 and BRCA1. Hum Mutat 24:272
Stella A, Wagner A, Shito K, Lipkin SM, Watson P, Guanti G, Lynch HT, Fodde R, Liu B (2001) A nonsense mutation in MLH1 causes exon skipping in three unrelated HNPCC families. Cancer Res 61:7020–7024
Vasen HF (2005) Clinical description of the Lynch Syndrome [Hereditary Nonpolyposis Colorectal Cancer (HNPCC)]. Fam Cancer 4(3):219–225
Wang Y, Friedl W, Lamberti C, Junck M, Mathiak M, Pagenstecher C, Propping P, Mangold E (2003) Hereditary nonpolyposis colorectal cancer: frequent occurrence of large genomic deletions in MSH2 and MLH1 genes. Int J Cancer 2003 Feb 20 103(5):636–641
Wehner M, Mangold E, Sengteller M, Friedrichs N, Aretz S, Friedl W, Propping P, Pagenstecher C (2005) Hereditary nonpolyposis colorectal cancer: pitfalls in deletion screening in MSH2 and MLH1 genes. Eur J Hum Genet 13:983–986
Acknowledgements
We would like to thank the patients who participated in this study and their attending doctors. The work of the consortium is supported by a multicenter grant from the Deutsche Krebshilfe, Bonn, Germany, project no. 70-3027-Ma1.
Author information
Authors and Affiliations
Corresponding author
Additional information
Databases: HNPCC – OMIM 114500, MSH2 – OMIM: 120435; GenBank: NM_000251.1, MLH1 – OMIM: 120436; GenBank: NM_000249.2, InSiGHT mutation database: http://www.insight-group.org/, Programs: BDGP: http://www.fruitfly.org/seq_tools/splice-instrucs.html, ESEfinder program: http://exon.cshl.edu/ESE/
Rights and permissions
About this article
Cite this article
Pagenstecher, C., Wehner, M., Friedl, W. et al. Aberrant splicing in MLH1 and MSH2 due to exonic and intronic variants. Hum Genet 119, 9–22 (2006). https://doi.org/10.1007/s00439-005-0107-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-005-0107-8