Abstract
CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR-associated protein) system is a crucial adaptive immune system for bacteria to resist foreign DNA infection. In this study, we investigated the prevalence and diversity of CRISPR/Cas systems in 175 Klebsiella oxytoca (K. oxytoca) strains. Specifically, 58.86% (103/175) of these strains possessed at least one confirmed CRISPR locus. Two CRISPR/Cas system types, I-F and IV-A3, were identified in 69 strains. Type I-F system was the most prevalent in this species, which correlated well with MLST. Differently, type IV-A3 system was randomly distributed. Moreover, the type IV-A3 system was separated into two subgroups, with subgroup-specific cas genes and repeat sequences. In addition, spacer origin analysis revealed that approximately one-fifth of type I-F spacers and one-third of type IV-A3 spacers had a significant match to MGEs. The phage tail tape measure protein and conjunctive transfer system protein were important targets of type I-F and IV-A3 systems in K. oxytoca, respectively. PAM sequences were inferred to be 5′-NCC-3′ for type I-F, 5′-AAG-3′ for subgroup IV-A3-a, and 5′-AAN-3′ for subgroup IV-A3-b. Collectively, our findings will shed light on the prevalence, diversity, and functional effects of the CRISPR/Cas system in K. oxytoca.






Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
Abbas AF, Al-Saadi AGM, Alkhudhairy MK (2020) Biofilm formation and virulence determinants of Klebsiella oxytoca clinical isolates from patients with colorectal cancer. J Gastrointest Cancer 51:855–860
Almendros C, Guzman NM, Garcia-Martinez J, Mojica FJ (2016) Anti-cas spacers in orphan CRISPR4 arrays prevent uptake of active CRISPR-Cas I-F systems. Nat Microbiol 1:16081
Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS (2016) PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16-21
Bai Z, Zhang S, Wang X, Aslam MZ, Wang W, Li H, Dong Q (2022) Genotyping based on CRISPR loci diversity and pathogenic potential of Diarrheagenic Escherichia coli. Front Microbiol 13:852662
Baker JL, Hendrickson EL, Tang X, Lux R, He X, Edlund A, McLean JS, Shi W (2019) Klebsiella and Providencia emerge as lone survivors following long-term starvation of oral microbiota. Proc Natl Acad Sci USA 116:8499–8504
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712
Biswas A, Gagnon JN, Brouns SJ, Fineran PC, Brown CM (2013) CRISPRTarget: bioinformatic prediction and analysis of crRNA targets. RNA Biol 10:817–827
Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964
Camargo AP, Nayfach S, Chen IA, Palaniappan K, Ratner A, Chu K, Ritter SJ, Reddy TBK, Mukherjee S, Schulz F, Call L, Neches RY, Woyke T, Ivanova NN, Eloe-Fadrosh EA, Kyrpides NC, Roux S (2023) IMG/VR v4: an expanded database of uncultivated virus genomes within a framework of extensive functional, taxonomic, and ecological metadata. Nucleic Acids Res 51:D733–D743
Carattoli A, Hasman H (2020) PlasmidFinder and in silico pMLST: identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol Biol 2075:285–294
Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Neron B, Rocha EPC, Vergnaud G, Gautheret D, Pourcel C (2018) CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res 46:W246–W251
Crowley VM, Catching A, Taylor HN, Borges AL, Metcalf J, Bondy-Denomy J, Jackson RN (2019) A type IV-A CRISPR-Cas system in Pseudomonas aeruginosa mediates RNA-guided plasmid interference in vivo. CRISPR J 2:434–440
Darby A, Lertpiriyapong K, Sarkar U, Seneviratne U, Park DS, Gamazon ER, Batchelder C, Cheung C, Buckley EM, Taylor NS, Shen Z, Tannenbaum SR, Wishnok JS, Fox JG (2014) Cytotoxic and pathogenic properties of Klebsiella oxytoca isolated from laboratory animals. PLoS One 9:e100542
Deveau H, Garneau JE, Moineau S (2010) CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64:475–493
Gagaletsios LA, Papagiannitsis CC, Petinaki E (2022) Prevalence and analysis of CRISPR/Cas systems in Pseudomonas aeruginosa isolates from Greece. Mol Genet Genom 297:1767–1776
Garcillan-Barcia MP, Francia MV, de la Cruz F (2009) The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev 33:657–687
Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71
Hogenauer C, Langner C, Beubler E, Lippe IT, Schicho R, Gorkiewicz G, Krause R, Gerstgrasser N, Krejs GJ, Hinterleitner TA (2006) Klebsiella oxytoca as a causative organism of antibiotic-associated hemorrhagic colitis. N Engl J Med 355:2418–2426
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169:5429–5433
Kamruzzaman M, Iredell JR (2019) CRISPR-Cas system in antibiotic resistance plasmids in Klebsiella pneumoniae. Front Microbiol 10:2934
Katsura I, Hendrix RW (1984) Length determination in bacteriophage lambda tails. Cell 39:691–698
Koonin EV, Makarova KS, Zhang F (2017) Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37:67–78
Krawczyk PS, Lipinski L, Dziembowski A (2018) PlasFlow: predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Res 46:e35
Kumar S, Stecher G, Peterson D, Tamura K (2012) MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 28:2685–2686
Leenay RT, Beisel CL (2017) Deciphering, communicating, and engineering the CRISPR PAM. J Mol Biol 429:177–191
Liao W, Liu Y, Chen C, Li J, Du F, Long D, Zhang W (2020) Distribution of CRISPR-cas systems in clinical carbapenem-resistant Klebsiella pneumoniae strains in a Chinese tertiary hospital and its potential relationship with virulence. Microb Drug Resist 26:630–636
Long J, Xu Y, Ou L, Yang H, Xi Y, Chen S, Duan G (2019) Diversity of CRISPR/Cas system in Clostridium perfringens. Mol Genet Genomics 294:1263–1275
Long J, Zhang J, Xi Y, Zhao J, Jin Y, Yang H, Chen S, Duan G (2023) Genomic insights into CRISPR-harboring plasmids in the Klebsiella genus: distribution, backbone structures, antibiotic resistance, and virulence determinant profiles. Antimicrob Agents Chemother 67:e0118922
Maiden MC, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND (2013) MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736
Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV (2011) Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477
Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV (2015) An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736
Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, Moineau S, Mojica FJM, Scott D, Shah SA, Siksnys V, Terns MP, Venclovas C, White MF, Yakunin AF, Yan W, Zhang F, Garrett RA, Backofen R, van der Oost J, Barrangou R, Koonin EV (2020) Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18:67–83
Martinez Arbas S, Narayanasamy S, Herold M, Lebrun LA, Hoopmann MR, Li S, Lam TJ, Kunath BJ, Hicks ND, Liu CM, Price LB, Laczny CC, Gillece JD, Schupp JM, Keim PS, Moritz RL, Faust K, Tang H, Ye Y, Skupin A, May P, Muller EEL, Wilmes P (2021) Roles of bacteriophages, plasmids and CRISPR immunity in microbial community dynamics revealed using time-series integrated meta-omics. Nat Microbiol 6:123–135
Mathers AJ, Cox HL, Kitchel B, Bonatti H, Brassinga AK, Carroll J, Scheld WM, Hazen KC, Sifri CD (2011) Molecular dissection of an outbreak of carbapenem-resistant enterobacteriaceae reveals Intergenus KPC carbapenemase transmission through a promiscuous plasmid. Mbio 2:e00204-00211
Mojica FJM, Diez-Villasenor C, Garcia-Martinez J, Almendros C (2009) Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology (reading) 155:733–740
Moradigaravand D, Martin V, Peacock SJ, Parkhill J (2017) Population structure of multidrug resistant Klebsiella oxytoca within hospitals across the UK and Ireland identifies sharing of virulence and resistance genes with K. pneumoniae. Genome Biol Evol 9:574–587
Neog N, Phukan U, Puzari M, Sharma M, Chetia P (2021) Klebsiella oxytoca and emerging nosocomial infections. Curr Microbiol 78:1115–1123
Netsvyetayeva I, Marusza W, Olszanski R, Szyller K, Krolak-Ulinska A, Swoboda-Kopec E, Sierdzinski J, Szymonski Z, Mlynarczyk G (2018) Skin bacterial flora as a potential risk factor predisposing to late bacterial infection after cross-linked hyaluronic acid gel augmentation. Infect Drug Resist 11:213–222
Newire E, Aydin A, Juma S, Enne VI, Roberts AP (2020) Identification of a type IV-A CRISPR-Cas system located exclusively on IncHI1B/IncFIB plasmids in Enterobacteriaceae. Front Microbiol 11:1937
Pinilla-Redondo R, Mayo-Munoz D, Russel J, Garrett RA, Randau L, Sorensen SJ, Shah SA (2020) Type IV CRISPR-Cas systems are highly diverse and involved in competition between plasmids. Nucleic Acids Res 48:2000–2012
Podschun R, Ullmann U (1998) Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603
Pourcel C, Touchon M, Villeriot N, Vernadet JP, Couvin D, Toffano-Nioche C, Vergnaud G (2020) CRISPRCasdb a successor of CRISPRdb containing CRISPR arrays and cas genes from complete genome sequences, and tools to download and query lists of repeats and spacers. Nucleic Acids Res 48:D535–D544
Russel J, Pinilla-Redondo R, Mayo-Munoz D, Shah SA, Sorensen SJ (2020) CRISPRCasTyper: automated identification, annotation, and classification of CRISPR-Cas loci. CRISPR J 3:462–469
Savino F, Cordisco L, Tarasco V, Calabrese R, Palumeri E, Matteuzzi D (2009) Molecular identification of coliform bacteria from colicky breastfed infants. Acta Paediatr 98:1582–1588
Shah SA, Erdmann S, Mojica FJ, Garrett RA (2013) Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol 10:891–899
Silas S, Lucas-Elio P, Jackson SA, Aroca-Crevillen A, Hansen LL, Fineran PC, Fire AZ, Sanchez-Amat A (2017) Type III CRISPR-Cas systems can provide redundancy to counteract viral escape from type I systems. Elife. https://doi.org/10.7554/eLife.27601
Sontheimer EJ, Marraffini LA (2010) Microbiology: slicer for DNA. Nature 468:45–46
Sorek R, Kunin V, Hugenholtz P (2008) CRISPR–a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol 6:181–186
Tse H, Gu Q, Sze KH, Chu IK, Kao RY, Lee KC, Lam CW, Yang D, Tai SS, Ke Y, Chan E, Chan WM, Dai J, Leung SP, Leung SY, Yuen KY (2017) A tricyclic pyrrolobenzodiazepine produced by Klebsiella oxytoca is associated with cytotoxicity in antibiotic-associated hemorrhagic colitis. J Biol Chem 292:19503–19520
van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ (2009) CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci 34:401–407
Westra ER, Buckling A, Fineran PC (2014) CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Microbiol 12:317–326
Yang L, Li W, Ujiroghene OJ, Yang Y, Lu J, Zhang S, Pang X, Lv J (2020) Occurrence and diversity of CRISPR Loci in Lactobacillus casei group. Front Microbiol 11:624
Yang J, Long H, Hu Y, Feng Y, McNally A, Zong Z (2022) Klebsiella oxytoca complex: update on taxonomy, antimicrobial resistance, and virulence. Clin Microbiol Rev 35:e0000621
Acknowledgements
The work was supported by the China Postdoctoral Science Foundation (2022M712859) and National Science and Technology Specific Projects (2018ZX10301407). This work was also supported by National Supercomputing Center in Zhengzhou.
Author information
Authors and Affiliations
Contributions
JL, CS and JZ designed the study. JL, YX and JF helped collected some data. JZ and JL analyzed data and wrote the paper. The paper was reviewed and edited by all of the authors. The final manuscript was read and approved by all authors.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Communicated by Martine Collart.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
438_2023_2065_MOESM1_ESM.eps
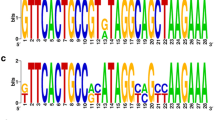
Supplementary file1 Figure S1 Repeat sequence alignment for type I-F (A), subgroups IV-A3-a (B), and IV-A3-b (C). The completely identical sequences are marked with dark blue, and the variations in nucleotide sites are marked with other colors. (EPS 1769 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, J., Xi, Y., Zhang, J. et al. Characterization and diversity of CRISPR/Cas systems in Klebsiella oxytoca. Mol Genet Genomics 298, 1407–1417 (2023). https://doi.org/10.1007/s00438-023-02065-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-023-02065-7