Abstract
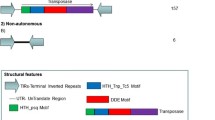
The availability of huge amounts of rice genome sequence now permits large-scale analysis of the structure and molecular characteristics of the previously identified transposase-encoding Rim2 (also called Hipa) element, which is transcriptionally activated by infection with the fungal pathogen Magnaporthe grisea and by treatment with the corresponding fungal elicitor. Based on genomic cloning and data mining from 230 Mb of rice genome sequence, 347 Rim2 elements, with an average size of 5.8 kb, were identified. This indicates that an estimated total of 600–700 Rim2 elements are present in the whole genome. Rim2 insertions occur non-randomly on the chromosomes, as visualized by fluorescence in situ hybridization. The elements harbor 16-bp terminal inverted repeats with the core sequence CACTG, 16-bp sub-terminal repeats, internal variable regions, 3-bp target sequence duplications in the flanking regions, and genes coding for Rim2 proteins (the putative transposase) and hydroxyproline-rich glycoproteins. High levels of insertion into genic regions are observed for members of this family, and the transposition history of the family can be deduced from the high level of shared sequences and analysis of repeat target sites of the elements. Phylogenetic analysis indicates that the putative RIM2 proteins fall into a subgroup distinct from the TNP2-like subgroup of transposases. Southern hybridization with genomic DNA from monocotyledonous and dicotyledonous plants demonstrates that the RIM2-coding sequence is unique to the Oryza genome. Our results demonstrate that the Rim2 elements from rice belong to a distinct superfamily of CACTA-like elements with evolutionary diversity.







Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Arnault C, Dufournel I (1994) Genome and stresses: reactions against aggressions, behavior of transposable elements. Genetica 93:149–160
Bercury SD, Panvas T, Irenze K, Walker EL (2001) Molecular analysis of the Doppia transposable element of maize. Plant Mol Biol 47:341–351
Bureau TE, Ronald PC, Wessler SR (1996) A computer-based systematic survey reveals the predominance of small inverted-repeat elements in wild-type rice genes. Proc Natl Acad Sci USA 93:8524–8529
Chopra S, Brendel V, Zhang J, Axtell JD, Peterson T (1999) Molecular characterization of a mutable pigmentation phenotype and isolation of the first active transposable element from Sorghum bicolor. Proc Natl Acad Sci USA 96:15330–15335
Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3:329–341
Gierl A (1996) The En/Spm transposable element of maize. Curr Topics Microbiol Immunol 204:145–159
Goff SA, et al (2002) A draft sequence of the rice genome ( Oryza sativa L. ssp. japonica). Science 296:92–100
Han CG, Frank MJ, Ohtsubo H, Ohtsubo E (2000) New transposable elements identified as insertions in rice transposon Tnr1. Genes Genet Syst 75:69–77
He ZH, Dong HT, Dong JX, Li DB, Ronald PC (2000) The rice Rim2 transcript accumulates in response to Magnaporthe grisea and its predicted protein product shares similarity with TNP2-like proteins encoded by CACTA transposons. Mol Gen Genet 264:2–10
Henikoff S, Henikoff JG (1993) Performance evaluation of amino acid substitution matrices. Proteins 17:49–61
Hirochika H (1993) Activation of tobacco retransposons during tissue culture. EMBO J 12:2521–2528
Hirochika H, Sugimito K, Otsuki Y, Tsugawa H, Kanda M (1996) Retrotransposon of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci USA 93:7783–7788
Jiang J, Gill BS, Wang GL, Ronald PC, Ward DC (1995) Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci USA 92:4487–4491
Jiang N, Wessler SR (2001) Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant Cell 13:2553–2564
Jiang N, Bao Z, Temnykh S, Cheng Z, Jiang J, Wing RA, McCouch SR, Wessler SR (2002) Dasheng. A recently amplified nonautonomous long terminal repeat element that is a major component of pericentromeric regions in rice. Genetics 161:1293–1305
Jiang N, Bao ZR, Zhang XY, Hirochika H, Eddy SR, McCouch S, Wessler S (2003) An active DNA transposon family in rice. Nature 421:163–167
Kapitonov VV, Jurka J (1999) Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica 107:27–37
Kidwell MG, Lisch D (1997) Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci USA 94:7704–7711
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245
Kunze R, Weil CF (2002) The hAT and CACTA superfamilies of plant transposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. ASM Press, Washington, pp 565–609
Kunze R, Saedler H, Lönnig WE (1997) Plant transposable elements. Adv Bot Res 27:331–470
Le QH, Wright S, Yu Z, Bureau T (2000) Transposon diversity in Arabidopsis thaliana. Proc Natl Acad Sci USA 97:7376–7381
Leprinc AS, Grandbastien MA, Christian M (2001) Retrotransposons of the Tnt1B family are mobile in Nicotiana plumbaginifolia and can induce alternative splicing of the host gene upon insertion. Plant Mol Biol 47:533–541
Mao L, Wood TC, Yu Y, Budiman MA, Tomkins J, Woo S, Sasinowski M, Presting G, Frisch D, Goff S, Dean RA, Wing RA (2000) Rice transposable elements: a survey of 73,000 sequence-tagged connectors. Genome Res 10:982–990
McClintock B (1984) The significance of responses of the genome to challenge. Science 226:792–801
McDonald JF (1995) Transposable elements:-possible catalysts of organismic evolution. Trends Ecol Evol 10:123–126
Melayah D, Bonnivard E, Chalhoub B, Audeon C, Grandbastien MA (2001) The mobility of the tobacco Tnt1 retrotransposon correlates with its transcriptional activation by fungal factors. Plant J 28:159–168
Mhiri C, Morel JB, Vernhettes S, Casacuberta JM, Lucas H, Grandbastien MA (1997) The promoter of the tobacco Tnt1 retrotransposon is induced by wounding and by abiotic stress. Plant Mol Biol 33:257–266
Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T (2001) Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411:212–214
Motohashi R, Ohtsubo E, Ohtsubo H (1996) Identification of Tnr3, a Suppressor-Mutator/Enhancer -like transposable element from rice. Mol Gen Genet 250:148–152
Nacken WKF, Piotrowiak R, Saedler H, Sommer H (1991) The transposable element Tam1 from Antirrhinum majus shows structural homology to the maize transposon En/Spm and has no sequence specificity of insertion. Mol Gen Genet 228:201–208
Ohtsubo H, Kumekawa N, Ohtsubo E (1999) RIRE2, a novel gypsy-type retrotransposon from rice. Genes Genet Syst 74:83–91
Ozeki Y, Davies E, Takeda J (1997) Somatic variation during long-term subculturing of plant cells caused by insertion of a transposable element in a phenylalanine ammonia-lyase (PAL) gene. Mol Gen Genet 254:407–416
Panaud O, Vitte C, Hivert J, Muzlak S, Talag J, Brar D, Sarr A (2002) Characterization of transposable elements in the genome of rice ( Oryza sativa L.) using representational difference analysis (RDA). Mol Genet Genomics 268:113–121
Panavas T, Weir J, Walker EL (1999) The structure and paramutagenicity of the R-marbled haplotype of Zea mays. Genetics 153:979–991
Pereira A, Cuyoers H, Gierl A, Schwarz-Sommer Z, Saedler H (1986) Molecular analysis of the En/Spm element system of Zea mays. EMBO J 5:835–841
Pouteau S, Boccara M, Grandbastien MA (1994) Microbial elicitors of plant defense responses activate transcription of a retrotransposon. Plant J 5:535–542
Rhodes PR, Vodkin LO (1988) Organization of the Tgm family of transposable elements in soybean. Genetics 120:597–604
Snowden KC, Napoli A (1998) PsI: a novel Spm -like transposable element from Petunia hybrida. Plant J 14:43–54
Song WY, Pi LY, Bureau TE, Ronald PC (1998) Identification and characterization of 14 transposon-like elements in the non-coding regions of the members of the Xa21 family of disease resistance genes in rice. Mol Gen Genet 258:449–456
Tarchini R, Biddle P, Wineland R, Tingey S, Rafalski A (2000) The complete sequence of 340 kb of DNA around the rice Adh1-Adh2 region reveals interrupted colinearity with maize chromosome 4. Plant Cell 12:381–391
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24:4876–4882
Turcotte K, Srinivasan S, Bureau T (2001) Survey of transposable elements from rice genomic sequences. Plant J 25:169–179
Vernhettes S, Grandbastien MA, Casacuberta JM (1997) In vivo characterization of transcriptional regulatory sequences involved in the defence-associated expression of the tobacco retrotransposon Tnt1. Plant Mol Biol 35:673–679
Wessler SR (1996) Plant retrotransposons: turned on by stress. Curr Biol 6:959–961
Witte CP, Le QH, Bureau T, Kumar A (2001) Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc Natl Acad Sci USA 98:13778–13783
Yu J, et al (2002) A draft sequence of the rice genome ( Oryza sativa L. ssp. indica). Science 296:79–92
Acknowledgements
We thank Pamela Ronald for providing the rice BACs, and Susan R. Wessler, Thomas Bureau and Antoni Rafalski for useful comments on this work. This work was funded by NSFC grants (30125030, 90208010), a CAS grant (KSCX2-SW-301-02) and a MOST of China grant (2001AA222321) to Z.H.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M.-A. Grandbastien
The first two authors contributed equally to this work
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Wang, GD., Tian, PF., Cheng, ZK. et al. Genomic characterization of Rim2 / Hipa elements reveals a CACTA-like transposon superfamily with unique features in the rice genome. Mol Genet Genomics 270, 234–242 (2003). https://doi.org/10.1007/s00438-003-0918-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-003-0918-z