Abstract
Introduction
Head and neck squamous cell carcinoma (HNSCC) is among the most common cancers in the world with a low survival rate and common diagnosis at late stages. Deubiquitination of proteins is involved in tumor growth, metastasis, apoptosis, and immunosuppressive pathways. The impact of the ubiquitin-specific protease (USP4) on survival was only scarcely investigated so far. The goal of our research was to analyze the association of USP4 expression with prognosis and clinicopathological features in HNSCC.
Methods
USP4 mRNA levels were derived from The Cancer Genome Atlas (TCGA) for a cohort of 510 patients. Protein expression of USP4 was analyzed by immunohistochemistry in a second cohort of 113 patients. Associations between USP4 levels and overall survival, disease-free survival and clinicopathological data were analyzed.
Results
High levels of USP4 mRNA were associated with prolonged overall survival in univariable analysis. There was no more association with survival after correction for the confounders HPV, stage and smoker status. High USP4 mRNA levels were linked to a lower T-stage, the patient’s age at diagnosis, and a positive HPV status. USP4 protein levels were not associated with prognosis or other features.
Conclusion
Since high USP4 mRNA was not an independent prognostic marker, we assume that the association is a result of the correlation of high USP4 mRNA with an HPV-positive status. Therefore, further investigation of USP4 mRNA and its association with the HPV status of HNSCC patients is warranted.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is one of the leading causes of cancer-associated death worldwide, with a high morbidity and only slight improvement of quality of life and life expectancy (Johnson et al. 2020). To reduce mortality and improve quality of life for HNSCC patients, treatment options need to be improved. In the recurrent and metastatic setting, the use of nivolumab and pembrolizumab have greatly improved therapeutic options (Ferris et al. 2016; Cohen et al. 2019; Burtness et al. 2019). However, overall survival remains low and the increase in survival rates is partly due to the emergence of HPV-positive HNSCC (Johnson et al. 2020; Chaturvedi et al. 2011). To date, only few prognostic factors have been identified (Panarese et al. 2019). Therefore, the aim of this study was to evaluate mRNA and protein expression of USP4 in HNSCC.
The ubiquitin–proteasome system is largely responsible for the balance between intracellular protein degradation and protein synthesis (He et al. 2017; Lim et al. 2013; Poondla et al. 2019). Aberrant expression of deubiquitinating enzymes (DUBs) has been implicated in the development of many pathologies. Particularly in cancer, malfunction of the ubiquitin–proteasome system has been linked to tumor formation and tumor metastasis. Crucial cancer-associated processes such as tumor growth, apoptosis, metastasis and chemoresistance are influenced by DUBs (Mennerich et al. 2019; Tanguturi et al. 2020; Poondla et al. 2019).
Ubiquitin-specific protease 4 (USP4) is a member of the DUB subfamily of cysteine proteases (Hu et al. 2021) and plays an essential physiological role by influencing DNA repair, cell growth, differentiation and immune response (S. Li et al. 2021). USP4 deubiquitinates a wide range of well-known target proteins thereby influencing important pathways such as TGF-beta, p53 or WNT/β-Catenin signaling (Hu et al. 2021; S. Li et al. 2021). Specifically, USP4 is indispensable for cellular homeostasis and balancing of the intracellular protein synthesis. Hence USP4 levels are strictly controlled and monitored under physiologic conditions (Hu et al. 2021). Frederick et al. were the first to note a reduced expression of USP4 in lung cancer relative to normal tissue samples (Frederick et al. 1998). Since then, alterations in USP4 expression were found in several cancer entities (Li et al. 2016; Xing et al. 2016; Wang et al. 2020; Zhong et al. 2018; Yao et al. 2017) including an upregulation in HNSCC (Hou et al. 2013). Additionally, USP4 is associated with outcome and progression of several cancers (Tao and You 2022; Zhou et al. 2019; Hu et al. 2021; Yao et al. 2017; Zhong et al. 2018; Wang et al. 2020).
Because of their potential to evolve tumor diagnosis and therapy, ubiquitin-specific proteases were studied extensively in the past (Vlasschaert et al. 2015). In particular, our previous work on PSMD14 (Schnoell et al. 2022), another member of the DUB system prompted us to investigate the role of USP4 in HNSCC. In this study, we aimed to elucidate USP4 mRNA and protein expression levels in relation to survival outcome and the association with clinicopathological aspects of HNSCC patients.
Patients and methods
The cancer genome atlas dataset (TCGA)
Patient data were drawn from cBioportal.org “The Cancer Genome Atlas (TCGA), Firehose Legacy” and supplemented with “TCGA, PanCancer Atlas” (Liu et al. 2018) and “TCGA, Nature 2015” (Lawrence et al. 2015) as described before by Schnoell et al. (2022). We included a total of 510 patients, who were treated between 1978 and 2012. Since perioperative complications could not be excluded, patients with an overall survival of less than two months were excluded from the panel, as were patients with incomplete survival records or missing mRNA data. The HPV status was determined by RNA sequencing of E6 and E7. Patients with over 1000 mapped RNA Seq reads were considered HPV positive (Liu et al. 2018). USP4 mRNA levels (RNA Seq V2 RSEM) were collected from cBioportal.org and classified as low or high expression based on a z-score > 0 criterion for high expression.
The tissue microarray (TMA) dataset
For the second patient cohort, a tissue microarray (TMA) database encompassing 113 HNSCC patients was probed for USP4 protein levels using immunohistochemistry. Included patients were diagnosed with HNSCC between 2002 and 2012 and all treated with surgery and post-operative radio(chemo)therapy. The HPV status was assessed by in situ hybridization. The TMA was generated by using a Galileo TMA CK Series-HTS Tissue computer assisted TMA Microarray platform (Integrated Systems Engineering Srl, Milan, Italy). Briefly, three cylindrical cores (2 mm in diameter) from formalin-fixed, paraffin-embedded HNSCC tissue were included from each patient. Conformation of histology was done by hematoxylin–eosin staining. The study was authorized by the Medical University of Vienna's ethical committee (EK1262/2019).
Immunohistochemistry
Immunohistochemistry was done following the manufacturer's protocol of the Ultra Vision Quanto Detection System HRP (Thermo Fisher Scientific, Waltham, MA, USA) with the following modifications (Schnoell et al. 2022). Briefly, samples were deparaffinized, rehydrated and incubated in H2O2 to suppress endogenous peroxidase activity. Samples were heated in EDTA in a microwave for antigen retrieval. Slides were then incubated with the Ultra V Block and afterwards with the USP4 primary antibody (1:500, HPA018499-25UL, Sigma Aldrich, Darmstadt, Germany) for 60 min at room temperature. Next, the primary antibody enhancer and horseradish peroxidase enhancer were applied. Stains were developed using DAB Quanto Detection System (Thermo Fisher Scientific, Waltham, MA, USA). Samples were washed with deionized water and counterstained with Hematoxylin Gill II (Merck, Darmstadt, Germany). Imaging was done with a PANNORAMIC Digital Slide Scanner and the open-source software QuPath (Edinburgh, Scotland). The h-score was calculated by combining the percentage of stained cells and the staining intensity (3 × % of cells with high positive staining + 2 × % of cells with moderately positive staining + 1 × % of cells with low positive staining). Staining was defined as high at a h-score cutoff value > 70 using the median h-score as reference.
Statistical analysis
STATA software (Stata Corp, Houston TX, USA, SCR_012763) and GraphPad Prism Software (GraphPad Prism Software, Inc., La Jolla, CA, USA, SCR_002798) were used to analyze all collected information. Continuous variables were evaluated with median values and 25th and 75th percentiles, while categorical data were reported in absolute and relative frequencies. The median observation was calculated as described by Schemper and Smith (1996). Overall survival and disease-free survival were calculated from time of initial pathological diagnosis until death or tumor recurrence, respectively. To visualize overall survival and disease-free survival, Kaplan–Meier curves were created. To evaluate the relation of clinicopathological features with USP4 expression, correlation analysis was performed using Fisher’s exact test and Chi2 test. Investigation of the association of USP4 mRNA and protein expression with overall survival and disease-free survival was done using log-rank test and uni- and multivariable analysis using the Cox proportional hazard regression model. A p value ≤ 0.05 was considered statistically significant.
Results
TCGA dataset: baseline characteristics
The TCGA dataset comprised 510 patients, 132 (26%) females and 378 (74%) males. Their characteristics are shown in Table 1. Median age at diagnose was 60.5 years (53–68). The majority of patients suffered from carcinoma of the oral cavity (60%), were diagnosed at stage IV (55%) and were HPV-negative (81%). The median observation time was 34.8 (21.6–55.2) months. The median overall survival was 56.9 (16.3–153.7) months, and the median disease-free survival was 56.4 (12.8–211.0) months.
TCGA dataset: association of USP4 mRNA levels with prognosis
Analysis of survival data is presented in Table 2 and Fig. 1. One-hundred and fifteen (23%) patients showed high USP4 mRNA expression levels. Patients with high USP4 mRNA expression showed improved overall survival outcomes compared to the low expression group (53.9 vs. 68.4 months; p = 0.036). In univariable analysis, patients with high USP4 expression were at lower risk of death (HR 0.68, 95% CI 0.48–0.98, p = 0.037). Subsequent evaluation of the continuous mRNA z-scores identified a reduced mortality risk for the group with high mRNA expression (HR 0.81, 95% CI 0.69–0.96, p = 0.013) in univariable analysis. Multivariable analysis, corrected for disease stage, smoker and HPV status, did not show an association of overall survival or disease-free survival with USP4 mRNA levels. Disease-free survival was not correlated to high USP4 mRNA expression (56.4 vs. 56.5 months; p = 0.170) nor the risk of recurrence (HR 0.78, 95% CI 0.55–1.11, p = 0.172; continuous: HR 0.88, 95% CI 0.75–1.04, p = 0.149).
The role of USP4 mRNA as a prognostic marker was further analyzed in the subgroups of HPV-positive and -negative patients. No association with overall or disease-free survival was found. Kaplan–Meier curves for the overall survival of patients with oropharyngeal carcinoma stratified by the HPV status and USP4 mRNA expression are shown in Fig. 2B.
Subgroup analysis of patients with oropharyngeal squamous cell carcinoma. A TCGA: USP4 mRNA expression (z-score) stratified by the HPV status. B TCGA: overall survival stratified by USP4 mRNA expression and HPV status. C TMA: USP4 protein expression (h-score) stratified by the HPV status. D TMA: overall survival stratified by USP4 mRNA expression and HPV status. HPV + : positive; HPV-: negative; USP4 + : high; USP4−: low
TCGA dataset: association of USP4 mRNA levels with clinicopathological data
To evaluate the association of USP4 mRNA expression with clinicopathological data, correlation analysis using Fisher’s exact or Chi2 test was performed (Table 3). High expression of USP4 mRNA was associated with lower age (< 60 years, p < 0.001), a lower T-stage (T1-2, p = 0.049) and an HPV-positive status (p < 0.001). The median USP4 z-score was 0.25 (− 0.49 to 1.10) in HPV-positive patients and − 0.71 (− 1.19to − 0.22) in HPV-negative patients. In oropharyngeal carcinoma patients the median z-score was 0.48 (− 0.31 to 1.37) in HPV-positive patients and − 0.68 (− 1.01to − 0.14) in HPV-negative patients (Fig. 2A). In oral carcinoma patients the median z-score was 0.10 (− 0.49 to 0.58) for HPV-positive patients and -0.76 (− 1.22 to − 0.27) for HPV-negative patients. The mRNA levels were not investigated separately for other locations of the primary due to the low number of HPV-positive patients.
TMA dataset: association of USP4 protein levels with outcome and clinicopathological data
A total of 113 patients, 27 females (24%) and 86 males (76%), at a median age of 59 (53–63) years were included in the TMA cohort. Most patients were HPV-negative (78%), nonsmokers (53%) and predominantly had a carcinoma of the oropharyngeal region (45%). Median observation time was 114.8 (76.3–141.7) months with a median overall survival time of 119.0 (30.72–not reached) months and a disease-free survival period of 156.8 (32.2–not reached) months. All patients were treated with surgery and post-operative radiotherapy.
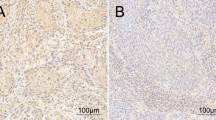
Protein expression was evaluated using immunohistochemistry and scored using the h-score. The median h-score of USP4 was 70.2 (47.4–82.8). The cutoff for high expression was > 70, using the median expression level as guidance. Thus, 57 patients (50%) showed high expression of USP4 demonstrated in Fig. 3. Kaplan–Meier curves were calculated (Fig. 1). Patients with high USP4 protein levels showed a trend for a prolonged overall survival (13.6 vs. 7.1, p = 0.598) and disease-free survival (not reached-12.7 months, p = 0.999). However, these results did not reach statistical significance. Further regression analysis did not show any association with the risk of death or recurrence in uni- or multivariable analysis (Table 2). There was no observed correlation with clinicopathological data (Table 3).
To further investigate the association of USP4 with the HPV status, subgroup analysis was performed. The median h-score for all HPV-positive patients was 68.5 (47.5–84.1) and 72.5 (47.3–83.0) for HPV-negative patients. In oropharyngeal cancer patients the median h-score was 68.6 (47.4–79.4) for HPV-positive patients and 78.8 (48.9–84.5) for HPV-negative patients (Fig. 2C). Due to the low number of HPV-positive patients, other locations of the primary tumor were not analyzed. Subgroup analysis of overall and disease-free survival of HPV-positive and -negative patients showed no association with outcome. Kaplan–Meier curves for the overall survival stratified by the HPV status und USP4 protein expression of oropharyngeal carcinoma patients are shown in Fig. 2D.
Discussion
HNSCC remains one of the most common carcinomas worldwide. The majority of patients are diagnosed at late disease stages with poor survival rates. As the needed multimodal therapy has many consequences and affects quality of life, better stratification of high and low risk patients would be beneficial. Thus, biomarkers are required to improve therapeutic decisions (Johnson et al. 2020). The deubiquitinating enzyme USP4 is associated with outcome in several carcinomas (Tao and You 2022; Zhou et al. 2019; Zhong et al. 2018; Wang et al. 2020; Yao et al. 2017). In HNSCC, USP4 is upregulated compared to non-cancerous tissue and has tumor suppressor qualities in vitro (Hou et al. 2013). However, the role of USP4 as a prognostic marker in HNSCC has not been investigated so far.
Our study aimed to investigate the association of USP4 protein and mRNA levels with HNSCC prognosis and clinicopathological data. First, mRNA expression data were derived from the TCGA dataset. Twenty-three percent of patients showed high mRNA expression. In survival analysis, high USP4 mRNA proved to be a marker for prolonged overall survival (53.9 vs. 68.4 months; log-rank p = 0.036) and a lower risk of death in univariable analysis (grouped: HR 0.68, 95% CI 0.48–0.98, p = 0.037, continuous: HR 0.81, 95% CI 0.69–0.96, p = 0.013). However, there was no more association with survival in multivariable analysis after correction for confounders (staging, HPV and smoker status). To further elucidate the change after multivariable analysis, we next investigated the correlation with clinicopathological data. This revealed associations of USP4 mRNA levels with T staging (p = 0.049) and HPV status (p < 0.001), but not with overall staging (p = 0.984) or smoker status (p = 0.982). HPV-negative patients show a worse outcome in oropharyngeal carcinoma (Paver et al. 2020), while the relevance for other sites is controversial (Tian et al. 2019; Zhu et al. 2019). Our survival analysis of the TCGA cohort showed that HPV was a positive predictive factor for a prolonged overall survival in the whole cohort (5.7 vs. 4.1 months, log-rank p = 0.002, univariable: HR 0.46, 95% CI 0.28–0.76 p = 0.002, multivariable adjusted for staging and smoker status: HR 0.43, 95% CI 0.26–0.72, p = 0.001). Therefore, we assume that the association of high USP4 mRNA levels with a better survival in the univariable analysis may be a result of the correlation of high USP4 mRNA levels with an HPV-positive status. In accordance with our results, Lohavanichbutr et al. showed that USP4 is downregulated in HPV-negative oropharyngeal cancer (Lohavanichbutr et al. 2009). Furthermore, the USP4 gene is an integration site for HPV16 in cervical and oropharyngeal squamous cell carcinoma, which may lead to alteration of gene expression (Díaz-Moreno et al. 2020). Hou et al. showed that USP4 has a tumor suppressor role in HNSCC in vitro (Hou et al. 2013). In other cancer entities, high USP4 mRNA was a positive prognostic marker in lung adenocarcinoma (Zhong et al. 2018) and a negative prognostic marker in glioblastoma patients (Zhou et al. 2019). Lung adenocarcinoma patients with a high USP4 status more commonly had no lymph node metastases (Zhong et al. 2018). However, in contrast to our results, USP4 prevailed as an independent prognostic factor for adenocarcinoma patients after correction for confounders in multivariable analysis of overall and disease-free survival (Zhong et al. 2018).
At protein level, the role of USP4 as a prognostic marker is controversial as well. High USP4 is associated with a worse outcome in gastric cancer (Tao and You 2022) and pancreatic cancer (Wang et al. 2020) and a better outcome in esophageal cancer (Yao et al. 2017). To elucidate whether the results of USP4 mRNA expression corresponded to protein levels in HNSCC patients, the association USP4 protein expression was analyzed in a secondary cohort. Tumor samples from a total of 113 patients (TMA dataset) were analyzed for USP4 protein levels semi-quantitively using immunohistochemistry. Patients with high USP4 protein levels showed a trend for an extended overall survival (13.6 vs. 7.1, p = 0.598) and disease-free survival (not reached vs. 12.7 months, p = 0.999). However, these results did not reach statistical significance. In cox regression analysis, USP4 protein levels (grouped and continuous) had no predictive power with respect to overall survival and or disease-free survival. Additional analysis of the association with clinicopathological data revealed no significant correlations. While it is reasonable to assume that mRNA and protein levels generally correlate with each other, post-translational mechanisms have the potential to alter protein expression (Liu et al. 2016). Furthermore, a selection bias may apply, since the second cohort (TMA) only contains patients who received surgery and post-operative radio(chemo)therapy, whereas the TCGA cohort also contains other treatment regimens.
Following additional limitations may have influenced the findings and conclusions of this study. A selection bias, leading to a skewed outcome might have been introduced through the retrospective study design. To minimize this risk, we included two separate cohorts. Although TMA cores were analyzed from several representative tumor areas, protein expression within other regions of the cancerous tissue might vary. To exclude type I and II errors due to a biased cutoff level of the categorical USP4 levels, we additionally analyzed the continuous z- and h-scores.
Altogether, this study showed that high USP4 mRNA is not an independent prognostic factor for a longer overall survival in HNSCC patients. The association with survival in univariable analysis may be due an association of high USP4 mRNA with HPV-positive patients. Therefore, further investigation of the role of USP4 in HPV-positive HNSCC is warranted. These findings were not validated in the second cohort at protein level.
Data availability
The datasets of this study are available from the corresponding author on reasonable request.
References
Burtness B, Harrington KJ, Greil R, Soulières D, Tahara M, de Castro G, Psyrri A et al (2019) Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. The Lancet 394(10212):1915–1928. https://doi.org/10.1016/S0140-6736(19)32591-7
Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang Bo et al (2011) Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 29(32):4294–4301. https://doi.org/10.1200/JCO.2011.36.4596
Cohen EEW, Soulières D, Le Tourneau C, Dinis J, Licitra L, Ahn MJ, Soria A et al (2019) Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. The Lancet 393(10167):156–167. https://doi.org/10.1016/S0140-6736(18)31999-8
Díaz-Moreno N, Osorio JC, García-Perdomo HA, Castillo A (2020) In silico analysis of genomic variables associated to HPV16 integration sites. Infectio 24(2):76
Ferris RL, Blumenschein G, Fayette J, Joel Guigay A, Colevas D, Licitra L, Harrington K et al (2016) Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med 375(19):1856–1867. https://doi.org/10.1056/NEJMoa1602252
Frederick A, Rolfe M, Isabel Chiu M (1998) The human UNP locus at 3p21.31 encodes two tissue-selective, cytoplasmic isoforms with deubiquitinating activity that have reduced expression in small cell lung carcinoma cell lines. Oncogene 16(2):153–165. https://doi.org/10.1038/sj.onc.1201537
He M, Zhou Z, George Wu, Chen Q, Wan Y (2017) Emerging role of DUBs in tumor metastasis and apoptosis: therapeutic implication. Pharmacol Ther 177(September):96–107. https://doi.org/10.1016/j.pharmthera.2017.03.001
Hou X, Wang L, Zhang L, Pan X, Zhao W (2013) Ubiquitin-specific protease 4 promotes TNF-α-induced apoptosis by deubiquitination of RIP1 in head and neck squamous cell carcinoma. FEBS Lett 587(4):311–316. https://doi.org/10.1016/j.febslet.2012.12.016
Hu B, Zhang D, Zhao K, Wang Y, Pei L, Qianmei Fu, Ma X (2021) Spotlight on USP4: structure, function, and regulation. Front Cell Dev Biol 9(February):1–15. https://doi.org/10.3389/fcell.2021.595159
Johnson DE, Barbara Burtness C, Leemans R, Lui VWY, Bauman JE, Grandis JR (2020) Head and neck squamous cell carcinoma. Nat Rev Dis Primers 6(1):92. https://doi.org/10.1038/s41572-020-00224-3
Lawrence MS, Sougnez C, Lichtenstein L, Cibulskis K, Lander E, Gabriel SB, Getz G et al (2015) Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517(7536):576–582. https://doi.org/10.1038/nature14129
Li Y, Jiang D, Zhang Qi, Liu X, Cai Z (2016) Ubiquitin-specific protease 4 inhibits breast cancer cell growth through the upregulation of PDCD4. Int J Mol Med 38(3):803–811. https://doi.org/10.3892/ijmm.2016.2685
Li S, Zhang H, Wei X (2021) Roles and mechanisms of deubiquitinases (DUBs) in breast cancer progression and targeted drug discovery. Life 11(9):965. https://doi.org/10.3390/life11090965
Lim K-H, Ramakrishna S, Baek K-H (2013) Molecular mechanisms and functions of cytokine-inducible deubiquitinating enzymes. Cytokine Growth Factor Rev 24(5):427–431. https://doi.org/10.1016/j.cytogfr.2013.05.007
Liu Y, Beyer A, Aebersold R (2016) On the dependency of cellular protein levels on MRNA abundance. Cell 165(3):535–550. https://doi.org/10.1016/j.cell.2016.03.014
Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ et al (2018) An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173(2):400-416.e11. https://doi.org/10.1016/j.cell.2018.02.052
Lohavanichbutr P, Houck J, Fan W, Yueh B, Mendez E, Futran N, Doody DR et al (2009) Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer. Arch Otolaryngol Head Neck Surg 135(2):180. https://doi.org/10.1001/archoto.2008.540
Mennerich D, Kubaichuk K, Kietzmann T (2019) DUBs, hypoxia, and cancer. Trends Cancer 5(10):632–653. https://doi.org/10.1016/j.trecan.2019.08.005
Panarese I, Aquino G, Ronchi A, Longo F, Montella M, Cozzolino I, Roccuzzo G, Colella G, Caraglia M, Franco R (2019) Oral and oropharyngeal squamous cell carcinoma: prognostic and predictive parameters in the etiopathogenetic route. Expert Rev Anticancer Ther 19(2):105–119. https://doi.org/10.1080/14737140.2019.1561288
Paver EC, Currie AM, Gupta R, Dahlstrom JE (2020) Human papilloma virus related squamous cell carcinomas of the head and neck: diagnosis, clinical implications and detection of HPV. Pathology 52(2):179–191. https://doi.org/10.1016/j.pathol.2019.10.008
Poondla N, Chandrasekaran AP, Kim K-S, Ramakrishna S (2019) Deubiquitinating enzymes as cancer biomarkers: new therapeutic opportunities? BMB Rep 52(3):181–189. https://doi.org/10.5483/BMBRep.2019.52.3.048
Schemper M, Smith TL (1996) A note on quantifying follow-up in studies of failure time. Control Clin Trials 17(4):343–346. https://doi.org/10.1016/0197-2456(96)00075-X
Schnoell J, Scheiflinger A, Al-Gboore S, Kadletz-Wanke L, Kenner L, Heiduschka G, Jank BJ (2022) The prognostic role of PSMD14 in Head and neck squamous cell carcinoma. J Cancer Res Clinical Oncol. https://doi.org/10.1007/s00432-022-04072-4
Tanguturi P, Kim K-S, Ramakrishna S (2020) The role of deubiquitinating enzymes in cancer drug resistance. Cancer Chemother Pharmacol 85(4):627–639. https://doi.org/10.1007/s00280-020-04046-8
Tao Y, You W (2022) The deubiquitinating enzyme USP4 functions as an oncoprotein in gastric cancer and mediates NF-ΚB signaling by regulating PRL-3 expression. Front Biosci Landm 27(10):286
Tian S, Switchenko JM, Jhaveri J, Cassidy RJ, Ferris MJ, Press RH, Pfister NT et al (2019) Survival outcomes by high-risk human papillomavirus status in nonoropharyngeal head and neck squamous cell carcinomas: a propensity-scored analysis of the national cancer data base. Cancer. https://doi.org/10.1002/cncr.32115
Vlasschaert C, Xia X, Coulombe J, Gray DA (2015) Evolution of the highly networked deubiquitinating enzymes USP4, USP15, and USP11. BMC Evol Biol 15(1):230. https://doi.org/10.1186/s12862-015-0511-1
Wang Y, Zhou L, Jun L, Jiang B, Liu C, Liang Z, Zhou W, Guo J (2020) Ubiquitin-specific protease 4 predicts an unfavorable prognosis and promotes malignant behaviors in vitro in pancreatic cancer. Exp Cell Res 396(2):112317. https://doi.org/10.1016/j.yexcr.2020.112317
Xing C, Xing-Xing Lu, Guo P-D, Shen T, Zhang S, He X-S, Gan W-J et al (2016) Ubiquitin-specific protease 4-mediated deubiquitination and stabilization of PRL-3 is required for potentiating colorectal oncogenesis. Can Res 76(1):83–95. https://doi.org/10.1158/0008-5472.CAN-14-3595
Yao R, Juan Pu, Fan R, Zhu W, Ding X, Shen X, Zhang T (2017) Ubiquitin-specific protease 4 improves the prognosis of the patients in esophageal cancer. Cancer Biomark 20(3):317–323. https://doi.org/10.3233/CBM-170308
Zhong M, Jiang Qi, Jin R (2018) USP4 expression independently predicts favorable survival in lung adenocarcinoma. IUBMB Life 70(7):670–677. https://doi.org/10.1002/iub.1755
Zhou Y, Liang P, Ji W, Zengpeng Yu, Chen H, Jiang Li (2019) Ubiquitin-specific protease 4 promotes glioblastoma multiforme via activating ERK pathway. Onco Targets Ther 12(March):1825–1839. https://doi.org/10.2147/OTT.S176582
Zhu Y, Xin X, Liming G, Xiaoli Z, Wenwen D, Zhiyong L, Zhiqiang G, Xingming C (2019) Prognostic implications of human papillomavirus type 16 status in non-oropharyngeal head and neck cancer: a propensity score matching analysis. Ann Transl Med 7(23):759–759. https://doi.org/10.21037/atm.2019.11.72
Funding
Open access funding provided by Medical University of Vienna. Not applicable.
Author information
Authors and Affiliations
Contributions
AS and JS contributed to the design of the study, statistical analysis, and writing of the manuscript. SA, BJJ, FB, LKW, LK, and GH contributed to the design of the study, data collection and manuscript revision. All authors critically reviewed the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This study was approved by the ethics committee of the Medical University of Vienna (EK1262/2019).
Consent for participation and publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Scheiflinger, A., Al-Gboore, S., Jank, B.J. et al. High USP4 mRNA is associated with an HPV-positive status in head and neck squamous cell carcinoma patients. J Cancer Res Clin Oncol 149, 10675–10683 (2023). https://doi.org/10.1007/s00432-023-04872-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-04872-2