Abstract
Introduction
Colorectal cancer (CRC) constitutes one of the most prevalent malignancies in the world. Recent research suggests that cancer stem cells (CSCs) are responsible for tumor cell’s malignant behavior in CRC. This study has been designed to determinate clinical implications of CSC markers: CD44, DCLK1, Lgr5, and ANXA2 in CRC.
Materials and methods
The study was performed on tissue samples which were collected from 89 patients undergoing colectomy. Formalin-fixed paraffin-embedded tissue blocks with representative tumor areas were identified and corded. Immunohistochemical staining was performed using anti-CD44, anti-LGR5, anti-ANXA2, and anti-DCLK1 antibodies. The H-score system was utilized to determine the immunointensity of CRC cells.
Results
The lower expression of Lgr5 was significantly correlated with the presence of lymph-node metastases (p = 0.011), while high expression of Lgr5 was statistically significant in vascular invasion in examined cancer tissue samples (p = 0.027). Moreover, a high H-score value of Lgr5 expression was significantly related to a reduced overall survival rate (p = 0.043).
Conclusion
Our results suggest a strong relationship between CSC marker Lgr5 and vascular invasion, presence of lymph-node metastasis, and overall poor survival. The presence of Lgr5 might be an unfavorable prognostic factor, and its high level in cancer tissue is related to an aggressive course. This marker could also be used to access the effectiveness of the treatment.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) constitutes one of the most prevalent malignancies in the world, contributing 9% of the total number of new cases diagnosed in 2018 in the United States (Siegel et al. 2019). Factors associated with an increased risk or development of CRC include obesity, physical inactivity, smoking, alcohol use, age, type 2 diabetes mellitus, and a family history of colon or rectal cancer (Marley and Nan 2016). It should be noted that CRC is a multifactorial disease, which is also connected with several genetic mutations such as changes in the APC, STK11, MYH, and mismatch repair genes (Genetics 2005). The staging system most often used for CRC is a classification system developed by the American Joint Committee on Cancer, which is based on three basic elements, first, the size and extent of the tumor (T), second, the number of nearby lymph nodes with cancer metastases (N), and, finally, distant metastases, for example, the spread of the primary tumor to other parts of the body (M). The 5-year survival rate for those diagnosed with early stage, localized disease (stages I and II) is approximately 90%; however, only about 39% of patients are diagnosed at this stage (Amin et al. 2017). Late-stage diagnosis (stages III and IV) of CRC is associated with a dramatically worse prognosis; the 5-year survival years fluctuate at around 13.1% (Simon 2016). Distant metastasis and recurrence are the major cause of patients' death, with more than 50% of CRC-related mortality are due to metastatic spread to the liver (Zarour et al. 2017).
Recent research suggests that cancer stem cells (CSCs) are responsible for tumor development and a tumor cell’s malignant behavior as well as metastasis in CRC (Ricci-Vitiani et al. 2009). CSCs are a subpopulation of cancer cells that have the ability of self-renewal, the potential to generate differentiated cells of the tissue of origin (multipotency), and resistance to chemotherapy, and have high tumorigenicity (Pang et al. 2010). Furthermore, it is well known that CSCs are insensitive to the current drug regimens. CSCs in CRC are identified via a group of surface markers, such as CD44, CD133, ANXA2, CD24, DCLK1, Lgr5, ALDH1, Nanog, Oct-4, SOX-2, and EpCAM (Leng et al. 2018) (Gzil et al. 2019a, b). Some of these markers could have a potential clinical role in predicting pathological stage, therapy resistance, and cancer recurrence in patients with colorectal carcinoma.
The current study has been designed to determinate clinical implications related to the expression level of chosen CSC markers of CRC and their importance in tumor progression and prognosis, which is still not well understood. The level of CSC markers such as CD44, DCLK1, Lgr5, and ANXA2 were investigated in correlation with clinical data and histopathological parameters. Moreover, to define the prognostic value of these proteins, the expression levels of investigated markers were compared with patient survival rate.
Materials and methods
Materials
The study was performed on tissue samples which were collected from 89 patients undergoing colectomy due to adenocarcinoma. The samples were excluded from further analysis; if patients received neoadjuvant chemotherapy before primary resection, the performed surgery was re-operated or resection of the recurrent tumor, and if the quality of the sample collected was unacceptable. The medical records include gender, age, pathological characteristic of resected material, and clinical outcome.
The investigated group consists of 37 female and 52 males. Excluded patients were diagnosed with an inherited predisposing condition for CRC development, such as familial adenomatous polyposis, hereditary nonpolyposis colorectal cancer, MUTYH-associated polyposis, or other polyposis syndromes. At the time of diagnosis, 18 patients suffered from type 2 diabetes mellitus, 9 were diagnosed with chronic kidney disease, and 26 had cardiovascular disease. Colorectal cancer manifested by anemia in 20 patients, and intestinal obstruction in 25 patients and in 6 perforation. In the case of 16 patients, first symptoms were related with the presence of distal metastasis, while 22 patients were diagnosed during screening program. Most common localizations of tumors were rectum (36%) and sigmoid colon (27%). Histopathological records revealed 16 cases (18%) with the invasion of muscularis propria (pT2), 58 cases (65%) with the invasion of subserosa or surrounding tissues (pT3), and 15 cases (17%) with the presence on visceral peritoneum or attached to neighboring structures (pT4). Metastases to regional lymph nodes were detected in 46 cases, while 43 patients (48%) had metastases to the liver (pM1a). 34 cases of metastases were present at the time of diagnosis (synchronous metastases), while 9 of the patients developed metastases after primary surgical treatment (metachronous metastasis). Median follow-up in investigated group amounted 19.8 months. The detailed clinical data of our groups are summarized in Table 1.
Methods
Tissue microarray construction
Formalin-fixed paraffin-embedded tissue blocks with representative tumor areas with at least 80% of tumor cells were identified through a review of corresponding hematoxylin–eosin (HE) stained slides. Areas of interest were identified and marked on each selected block. The tissue microarrays paraffin block was cored using a 2-mm core. For each case, two or three representative cores of tumor were arrayed. The cores were transferred to the “donor block” using an automated tissue arrayer (TMA Master; 3DHISTECH, Budapest, Hungary). Next, paraffin-embedded TMA block was cut into 3–4 μm-thick sections, using a manual rotary microtome (Accu-Cut, Sakura Finetek, Torrance, CA, USA). The prepared sections were then placed on extra adhesive slides (Superfrost Plus; Menzel-Glaser, Braunschweig, Germany).
Immunohistochemical staining was performed using DakoAutostainer Link 48 (Dako, Agilent Technologies, USA) and BenchMark® Ultra automated slide processing system (Ventana Medical Systems, Tucson, AZ, USA). The following primary antibodies were used: rabbit monoclonal anti-CD44 (SP37) antibody (ready to use, Ventana Medical Systems, Tucson, AZ, USA), rabbit polyclonal anti-DCLK1 antibody (HPA015655, Sigma-Aldrich, Merck KGaA, Darmstadt, Germany), rabbit polyclonal anti-LGR5 antibody (HPA012530, Sigma-Aldrich, Merck KGaA, Darmstadt, Germany), and rabbit polyclonal anti-ANXA2 antibody (HPA046964, Sigma-Aldrich, Merck KGaA, Darmstadt, Germany). In the beginning, standardization and optimization of the IHC method were performed on a tissue recommended based on the antibody datasheet and reference sources (The Human Protein Atlas: https://www.proteinatlas.org) (Uhlen et al. 2010).
Immunohistochemical staining of LGR5, ANXA2, and DCLK1
Immunohistochemical staining of anti-LGR5, anti-ANXA2, and anti-DCLK1 antibodies was done using the DakoAutostainer Link 48 and Dako PT Link pre-treatment module. Prepared slides with tissue sections were deparaffinized and rehydrated. In the first step, the slides were heated in a high-pH buffer (Dako, Agilent Technologies, USA) at 95–98 °C for 20 min in PT Link (Dako, USA) for antigen retrieval. Then, the endogenous peroxidase activity was inhibited using 3% H2O2 for 10 min at room temperature (RT). The slides were treated with 3% bovine serum albumin solution for 15 min at RT to block non-specific antibody binding sites. Next, the sections were incubated with rabbit polyclonal anti-DCLK1 antibody (1:100), rabbit polyclonal anti-LGR5 antibody (1:500), and rabbit polyclonal anti-ANXA2 antibody (1:100) for 30 min. Subsequently, slides washed three times with phosphate-buffered saline. After adding the secondary horseradish peroxidase (HRP, Dako, Agilent Technologies) labeled antibody for 20 min at RT, the 3,3′diaminobenzidine (DAB) was used to detect the localization of the antigen–antibody complex. The sections were counterstained in hematoxylin and washed. Finally, tissue sections were dehydrated in increasing ethanol concentrations (80, 90, 96, and 99.8%), cleared in xylenes (I–IV), mounted using mounting medium, and observed.
Immunohistochemical staining of CD44
Immunohistochemical staining of anti-CD44 was done using the BenchMark® Ultra automated slide processing system (Ventana Medical Systems, Tucson, AZ, USA). In the first step, deparaffinization and rehydration were performed in EZ Prep solution (Ventana Medical Systems, Tucson, AZ, USA). Next, antigen retrieval of tissue sections was performed in Cell Conditioning (CC2) solution for 68 min. Incubation with the primary rabbit monoclonal anti-CD44 (SP37) antibody was performed for 20 min at 36 °C. The reaction was performed using the visualization system (UltraView DAB Detection Kit; Ventana Medical Systems, Tucson, AZ, USA). The slides were counterstained with Hematoxylin II for 12 min and Bluing Reagent for 4 min. Finally, tissue sections were dehydrated in increasing ethanol concentrations (80, 90, 96, and 99.8%), cleared in xylenes (I–IV), mounted using mounting medium, and observed.
Evaluation of immunohistochemistry staining
The pathologists who were evaluating the immunohistochemical expression of examined antigens worked independently, and they were blinded from the patients’ clinical, as well as other data. The protein expression was evaluated using light microscope ECLIPSE E800 (Nikon Instruments Europe, Amsterdam, Netherlands) at 20× original objective magnification. The staining intensity was measured on a four-point scale as negative (0), week (1), moderated (2), and strong (3). The percentage of IHC positive cells were also recorded. The H-score was assigned due to calculation of the percentage of cells at each staining intensity level according to the following formula: H-score = [1 × (% cells 1 +) + 2 × (% cells 2 +) + 3 × (% cells 3 +)]. The H-score amounted maximal 300 and minimal 0 points (Yeo et al. 2015).
Statistical analysis
All statistical analyses were performed using Statistica version 13 (StatSoft) and Microsoft Excel 2007. The expression values of analyzed proteins were presented 25th percentile (25p), the median (M), and the 75th percentile (75p). The comparative studies were analyzed statistically using the nonparametric U Mann–Whitney and Kruskal–Wallis test. Overall survival (OS) curves were designed using ROC curves and performed with the Kaplan–Meier method and compared with the log-rank test. The p value < 0.05 was considered statistically significant.
Results
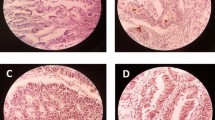
In the current study, there was observed the nucleocytoplasmic expression of Lgr5, DCLK1, and ANXA2 (Fig. 1) and membranous expression of CD44 had. The expression of Lgr5 was found in 92% of samples, DCLK1 in 97% cases, ANXA2 in 88% cases, and CD44 in 82% cases (Tables 2, 3). In most of the cases, a co-expression of all investigated CSC markers was detected (Table 4). Additionally, the presence of stromal markers LGR5, CD44, ANXA2, and DCLK1 staining has been identified as a common finding among adjacent microenvironment of the CRC.
Representative images of cancer stem cell markers by immunohistochemistry in colorectal cancer tissue with a low expression of Lgr5, b high expression of Lgr5, c low expression of DCLK1, d high expression of DCLK1, e low expression of ANAX2, f high expression of ANAX2, g low expression of CD44, and h high expression of CD44
Statistical analysis showed significant differences in expression of Lgr5 and the presence of lymph-node metastases (p = 0.011). The study revealed a lower expression of Lgr5 in both pN1 and pN2 status compared to pN0 (Fig. 2). Moreover, higher Lgr5 expression was statistically significantly related to vascular invasion in examined cancer tissue samples (p = 0.027; Fig. 3). However, statistical analysis did not show any correlations between Lgr5 and tumor localization, grade, T stage, the presence of distant metastases, and infiltration of neural structures. The remaining investigated CSC markers (DCLK1, ANXA2, and CD44) did not demonstrate a statistically significant relationship with tested clinicopathological features of colorectal cancer. All results are shown in Table 5.
The ROC curves were performed to receive the optimal cut-off points for the expression level of all investigated CSC markers. The expressions levels of Lgr5, DCLK1, ANXA2, and CD44 were divided into two groups, the first with a high expression and the second with a low expression. The obtained results were analyzed in correlation with an OS after a primary surgery by use of Kaplan–Meier survival curves (Fig. 4). In the case of Lgr5, a high H-score value was significantly related to reduced OS rate (p = 0.043). The remaining proteins did not demonstrate a statistically significant correlation.
Discussion
The current study aimed to investigate the expression of commonly occurring cancer stem cell markers of colorectal cancer. Our study showed that increased Lgr5 expression is statistically significantly related to a lack of lymph-node metastasis, presence of vascular invasion, and low survival rate in CRC. Moreover, the determination of CSC expression in postoperative tissue could be used in clinical practice as a prognostic factor but also to help with treatment planning.
Lgr5+ cells have been frequently investigated as CSC in colorectal cancer (Shimokawa et al. 2017). The expression of Lgr5 occurs between 56.3% and 82.4% in comparison to between 6 and 25% of Lgr5+ cells among crypt cells of the normal intestine (Ziskin et al. 2013; He et al. 2014; Jia et al. 2015; Zheng et al. 2018; Wang et al. 2018b). Furthermore, the series of studies have suggested that enhancement of Lgr5 expression is correlated with an evolution from adenoma to adenocarcinoma. (Fan et al. 2010; Zheng et al. 2018). In our study, 92% of exanimated colorectal specimens were Lgr5+ . The overexpression of Lgr5 in the colorectal cancer cells occurs due to up or downregulation of several noncoding RNAs, such as CASC15 miR-4310 or miR-23a, as a consequence of activation of PI3K/Akt signaling pathway (Takahashi et al. 2011; Mukohyama et al. 2017). Other studies have also emphasized the role of EGF/EGFR/STAT3/PDGFA pathway and histone acetylation on the Lgr5 promoter (Cheng et al. 2017, 2018).
From the previous studies on CRC, it is known that Lgr5 enhances downstream Wnt/β-catenin signaling and Lgr5+ cells exhibit a high ability of colony formation, self-renewal, differentiation, and Ki67 proliferative index (Fan et al. 2010; Takahashi et al. 2011; He et al. 2014; Leng et al. 2018; Zheng et al. 2018; Salehizadeh et al. 2019). The recent works have emphasized that a high level of this CSC marker is positively correlated with regards to both tumor size and the depth of invasion into surrounding tissues. (Fan et al. 2010; Hsu et al. 2013; Jiang et al. 2015; Nishioka et al. 2018; Zheng et al. 2018). Zhou et al. observed in their study that patients with CRC had reduced Lgr5 expression in more advanced stages (Zhou et al. 2017). However, our result did not show any correlation between Lgr5 and primary tumor size (T stage), which is supported by He et al.’s study, with a similar conclusion to our finding (He et al. 2014). A complete explanation of this issue needs more research; nevertheless, our results suggest a different role of Lgr5 in cancer development.
Moreover, some studies suggest that the degree of cancer differentiation (grading) is also highly associated with Lgr5 expression (Wu et al. 2012; Wang et al. 2018b). However, a couple of studies have demonstrated a low level of Lgr5+ cells in high grade, poorly differentiated CRC (Dame et al. 2018; Sato et al. 2019). In our study, a statistically significant correlation between histological grade and Lgr5 expression was not observed. Similar lack of relationship was also observed by other authors in their studies (He et al. 2014; Jiang et al. 2015).
The association Lgr5 and metastatic ability of CRC seem to be even more controversial than the above-discussed parameters. Our results suggest that the expression of Lgr5 is significantly lower in CRC with lymph-node metastases. However, the majority of until now published studies reported the opposite relationship between these features in CRC (Wu et al. 2012; He et al. 2014; Jiang et al. 2015; Zheng et al. 2018; Wang et al. 2018b). Most of the authors explained that the main roles of CSC are to migrate to the target tissue and form secondary tumors. The recent advancement in molecular biology seems to give the indications to understanding the underlying molecular pathways, which could make our result more reliable. Studies have shown that overexpression of Lgr5 increases cells adhesion due to enhance levels of cortical F-actin (Carmon et al. 2017; Walker et al. 2011). On the other hand, Lgr5- subpopulation of CRC CSC seems to be characterized by a higher expression of mesenchymal-associated genes such as Snail, Slug, Zeb1 and 2, and N-cadherin, and by a lower expression of epithelial-associated genes such as E-cadherin, occluding, and epithelial cell adhesion molecule (EpCAM) (Leng et al. 2018). According to some studies, these events could be explained by the fact that the sphere formation of CSC (and possible EMT) is associated with a decreased Lgr5 expression due to progressive CpG island methylation of its promoter during the progression of tumorigenesis (Jang et al. 2018). To summarize, due to downregulation of Lgr5 expression, intracellular mechanisms promoting cell adhesion may decrease with EMT promotion, leading to an increased metastatic ability of CSC. This mechanism seems to explain why a low expression of Lgr5 in a primary tumor may correlate with the presence of lymph-node metastases observed in the current study (Fig. 5). However, our results did not confirm any correlation between Lgr5 expression and distal metastasis.
The cell adhesion is increased due to enhance levels of cortical F-actin in colorectal cancer cells with overexpression of Lgr5. The colorectal cancer cells could change their phenotype into mesenchymal-like with high expression of Snail, Slug, Zeb1 and 2, and N-cadherin and low expression of epithelial-associated genes such as E-cadherin after the downregulation of Lgr5 due to progressive CpG island methylation, what lead to epithelial–mesenchymal transition and induction of metastatic process (Walker et al. 2011; Carmon et al. 2017; Jang et al. 2018; Leng et al. 2018). EpCAM—epithelial cell adhesion molecule; Lgr5—leucine-rich repeat-containing G-protein-coupled receptor 5; ZEB1—zinc-finger E-box-binding homeobox 1; ZEB2—zinc-finger E-box-binding homeobox 2; Snail—zinc-finger protein SNAI; Snug—zinc-finger protein SNAI2; EMT—epithelial–mesenchymal transition
Our analysis of Lgr5 expression and OS showed that a high level of Lgr5 could decrease the patient’s overall survival time. This remark suggests that Lgr5 could be used in clinical practice as a potential prognostic factor. A similar conclusion of a high level of Lgr5 is an unfavorable factor with a shorter OS and disease-free survival (independent from tumor size) was also given in many other studies (Takahashi et al. 2011; Wu et al. 2012, 2016; Hsu et al. 2013; Jiang et al. 2015; Nishioka et al. 2018). There are a couple of possibilities that could explain this observation. First, our study revealed the high expression of Lgr5 to be correlated with vascular invasion, which may indirectly indicate the presence of some unknown mechanism that may contribute to the ability of cancer cells to invade the vessels. It has been shown that Lgr5 enhances downstream Wnt/β-catenin signaling and consequently deregulation of factors such as c-MYC or CDKN1A. (Fan et al. 2010; Takahashi et al. 2011; He et al. 2014; Zheng et al. 2018). WNT signaling cascades in Lgr5+ cell cross-talk with other pro-tumorigenic components, for example, the FGF, Notch, Hedgehog, and TGFβ/BMP signaling, and promotes EMT of Lgr5+ cells (Wu et al. 2016; Jang et al. 2018; Solomon et al. 2018; Zhang et al. 2018). Second, CSCs are a subpopulation of cancer cells that are resistant to chemotherapy. Several studies suggested Lgr5+ as resistant not only to 5-FU but also to other components of commonly used CRC chemotherapy regimens such as Oxaliplatin and Irinotecan (Kobayashi et al. 2012; Zhang et al. 2019). Balancing both observations, it might be possible to deduce that high subpopulation of Lgr5+ cells due to increased spread of cancer cells including CSC into bloodstream creates a CRC cellular pool, which survives resection of the primary tumor, and is then responsible for resistance to adjuvant chemotherapy. Furthermore, conversion of LGR5+ to LGR5- CSC has been shown to enhance drug resistance in CRC (Hou et al. 2018). It could be possible that the subpopulation of Lgr5+ CSC grow during cancer progression and represent CRC cells, which could change into chemo-resistant Lgr5- cells with metastatic ability during cancer progression. This suggestion could explain why a weak expression of Lgr5 in CRC correlate with lymph-node metastasis (high population of present Lgr5- CSC at the time of diagnosis) and its high level with worse overall survival (the detection of all cells, which might transform into metastatic-related chemo-insensible clone). However, Wang et al. suggested the only nonsignificant relationship of Lgr5 alone with OS and Lgr5 has been in addition described in Jiang et al.’s study, different from our results as prognostic marker for better clinical outcome in CRC patients (Wang et al. 2018a; Zheng et al. 2018).
Our study did not show any significant correlation between CD44, DCLK1, and ANXA2 expression and clinicopathological features of colorectal cancer and OS. Recent studies have also demonstrated no significant association between CD44 and tumoral characteristics of CRC (Rohani et al. 2017; Wang et al. 2019). However, some studies have shown that CD44 overexpression has been correlated with worse OS of patients with CRC (Wang et al. 2019,2017). Similarly, several studies show a positive correlation between high DCLK1 expression and CRC clinicopathological characteristics and poor OS (GZIL et al. 2019a, b). However, Dai T. et al.’s study reveals that a high DCLK1 expression in CRC tissue could play a protective role against tumor progression and correlates with longer survival time (Dai et al. 2018). Furthermore, recent studies suggested a statistically significant correlation between high ANXA2 protein expression and histological grade, pTNM stage, and worse OS than in patients with low ANXA2 (Duncan et al. 2008; Yang et al. 2013).
Although it was not the aim of our study, we observed the expression of all investigated CSC markers in the stroma of CRC samples. Previous studies are consistent with our findings. An earlier study suggested that stromal staining of LGR5 could be associated with cancer with an advanced stage of CRC (Dame et al. 2018). Others have identified the presence of Lgr5+ stromal cells in the oral mucosa and in lung alveolar mesenchyme (Boddupally et al. 2016; Lee et al. 2017). In case of CD44, this protein is a cell surface glycoprotein expressed commonly on lymphocytes, monocytes, and granulocytes and it has been interestingly suggested, that the absence of stromal CD44 expression was connected with an increased death rate of patients with CRC (Furuta et al. 1996, 1998; Cairns et al. 2001). Latest studies highlighted the role of ANXA2 expression in cancer-associated fibroblasts in stromal tissue in pancreatic ductal carcinoma and epithelial ovarian cancer as a predictive biomarker for overall survival (Paliwal et al. 2012; Christensen et al. 2019). Other studies indicate that DCLK1 has been shown to be expressed in the stroma of colon, pancreatic, prostate, breast cancers, esophageal adenocarcinoma, and non-small cell lung cancer (NSCLC) (Sureban et al. 2011; Whorton et al. 2015; Tao et al. 2017). Moreover, Tao A. et al.’s study reveals association between DCLK1 expression in NSCLC cells and the tumor stroma and their correlation with worse prognosis (Tao et al. 2017). Future studies should focus on complex understanding of the significance of CSC markers-expressing stromal cells in colorectal carcinogenesis.
In conclusion, our results suggested a strong relationship between CSC marker Lgr5 and vascular invasion, presence of lymph-node metastasis, and poor survival. Our study suggests Lgr5 as an unfavorable prognostic factor and its high level in CRC could direct to a more precise classification of treatment and observation in patients with a shorter survival life-span. Dualistic nature of Lgr5+ CSC, of which decreased expression is correlated with nodal spread and increased with vascular invasion and shorter overall survival, needs to be more investigated, because our results in combination with earlier molecular biology studies suggest interesting nature of those cells.
Data availability
Author decelerates that all data and material are available by corresponding author.
References
Amin MB, Greene FL, Edge SB et al (2017) The Eighth Edition AJCC Cancer Staging Manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. https://doi.org/10.3322/caac.21388
Boddupally K, Wang G, Chen Y, Kobielak A (2016) Lgr5 marks neural crest derived multipotent oral stromal stem cells. Stem Cells 34:720–731. https://doi.org/10.1002/stem.2314
Cairns AP, Crockard AD, McConnell JR et al (2001) Reduced expression of CD44 on monocytes and neutrophils in systemic lupus erythematosus: relations with apoptotic neutrophils and disease activity. Ann Rheum Dis 60:950–955. https://doi.org/10.1136/ard.60.10.950
Carmon KS, Gong X, Yi J et al (2017) LGR5 receptor promotes cell-cell adhesion in stem cells and colon cancer cells via the IQGAP1-Rac1 pathway. J Biol Chem 292:14989–15001. https://doi.org/10.1074/jbc.M117.786798
Cheng CC, Chang J, Huang SCC et al (2017) YM155 as an inhibitor of cancer stemness simultaneously inhibits autophosphorylation of epidermal growth factor receptor and G9a-mediated stemness in lung cancer cells. PLoS ONE. https://doi.org/10.1371/journal.pone.0182149
Cheng C-C, Liao P-N, Ho A-S et al (2018) STAT3 exacerbates survival of cancer stem-like tumorspheres in EGFR-positive colorectal cancers: RNAseq analysis and therapeutic screening. J Biomed Sci 25:60. https://doi.org/10.1186/s12929-018-0456-y
Christensen MV, Høgdall C, Jensen SG et al (2019) Annexin A2 and S100A10 as candidate prognostic markers in epithelial ovarian cancer. Anticancer Res 39:2475–2482. https://doi.org/10.21873/anticanres.13367
Dai T, Hu Y, Lv F et al (2018) Analysis of the clinical significance of DCLK1+ colorectal cancer using novel monoclonal antibodies against DCLK1. Onco Targets Ther 11:5047–5057. https://doi.org/10.2147/OTT.S169928
Dame MK, Attili D, McClintock SD et al (2018) Identification, isolation and characterization of human LGR5-positive colon adenoma cells. Development. https://doi.org/10.1242/dev.153049
Duncan R, Carpenter B, Main LC et al (2008) Characterisation and protein expression profiling of annexins in colorectal cancer. Br J Cancer 98:426–433. https://doi.org/10.1038/sj.bjc.6604128
Fan X-S, Wu H-Y, Yu H-P et al (2010) Expression of Lgr5 in human colorectal carcinogenesis and its potential correlation with beta-catenin. Int J Colorectal Dis 25:583–590. https://doi.org/10.1007/s00384-010-0903-z
Furuta K, Zahurak M, Yang XL et al (1996) Relationship between CD44 expression and cell proliferation in epithelium and stroma of colorectal neoplasms. Am J Pathol 149(4):1147–1155
Furuta K, Zahurak M, Goodman SN et al (1998) CD44 expression in the stromal matrix of colorectal cancer: association with prognosis. Clin Cancer Res 4:21–29
Genetics M (2005) Lynch syndrome genes. Fam Cancer 63:227–232. https://doi.org/10.1007/s10689-004-7993-0
Gzil A, Szylberg Ł, Jaworski D et al (2019a) The essential role of DCLK1 in pathogenesis, diagnostic procedures and prognostic stratification of colorectal cancer. Anticancer Res 39:2689–2697
Gzil A, Szylberg Ł, Jaworski D et al (2019b) The essential role of DCLK1 in pathogenesis, diagnostic procedures and prognostic stratification of colorectal cancer. Anticancer Res 39:2689–2697. https://doi.org/10.21873/anticanres.13394
He S, Zhou H, Zhu X et al (2014) Expression of Lgr5, a marker of intestinal stem cells, in colorectal cancer and its clinicopathological significance. Biomed Pharmacother 68:507–513. https://doi.org/10.1016/j.biopha.2014.03.016
Hou MF, Chen PM, Chu PY (2018) LGR5 overexpression confers poor relapse-free survival in breast cancer patients. BMC Cancer. https://doi.org/10.1186/s12885-018-4018-1
Hsu H-C, Liu Y-S, Tseng K-C et al (2013) Overexpression of Lgr5 correlates with resistance to 5-FU-based chemotherapy in colorectal cancer. Int J Colorectal Dis 28:1535–1546. https://doi.org/10.1007/s00384-013-1721-x
Jang BG, Kim HS, Chang WY et al (2018) Expression profile of LGR5 and its prognostic significance in colorectal cancer progression. Am J Pathol 188:2236–2250. https://doi.org/10.1016/j.ajpath.2018.06.012
Jia H, Xiang L, Wang Z, Zhou Q (2015) A study on the mechanism of low-expressed cancer stem cell marker Lgr5 in inhibition of the proliferation and invasion of colorectal carcinoma. Cell Biochem Biophys 73:393–397. https://doi.org/10.1007/s12013-015-0640-6
Jiang Y, Li W, He X et al (2015) Lgr5 expression is a valuable prognostic factor for colorectal cancer: evidence from a meta-analysis. BMC Cancer. https://doi.org/10.1186/s12885-015-1985-3
Kobayashi S, Yamada-Okabe H, Suzuki M et al (2012) LGR5-positive colon cancer stem cells interconvert with drug-resistant LGR5-negative cells and are capable of tumor reconstitution. Stem Cells 30:2631–2644. https://doi.org/10.1002/stem.1257
Lee JH, Tammela T, Hofree M et al (2017) Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell 170:1149–1163.e12. https://doi.org/10.1016/j.cell.2017.07.028
Leng Z, Xia Q, Chen J et al (2018) Lgr5+CD44+EpCAM+ strictly defines cancer stem cells in human colorectal cancer. Cell Physiol Biochem 46:860–872. https://doi.org/10.1159/000488743
Marley AR, Nan H (2016) Epidemiology of colorectal cancer. Int J Mol Epidemiol Genet
Mukohyama J, Shimono Y, Minami H et al (2017) Roles of microRNAs and RNA-binding proteins in the regulation of colorectal cancer stem cells. Cancers (Basel). https://doi.org/10.3390/cancers9100143
Nishioka M, Suehiro Y, Sakai K et al (2018) TROY is a promising prognostic biomarker in patients with colorectal cancer. Oncol Lett 15:5989–5994. https://doi.org/10.3892/ol.2018.8079
Paliwal VK, Rahi SK, Singh P, Gupta RK (2012) “Wine-glass appearance” of pyramidal tracts in a patient with primary lateral sclerosis. Neurol India 60:110–111. https://doi.org/10.4103/0028-3886.93606
Pang R, Law WL, Chu ACY et al (2010) A subpopulation of CD26+cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell. https://doi.org/10.1016/j.stem.2010.04.001
Ricci-Vitiani L, Fabrizi E, Palio E, De Maria R (2009) Colon cancer stem cells. J Mol Med 87:1097–1104. https://doi.org/10.1007/s00109-009-0518-4
Rohani P, Noroozinia F, Modarresi P, Abbasi A (2017) CD44 standard isoform; not a good marker for colon cancer. Int J Cancer Manag. https://doi.org/10.5812/ijcm.9166
Salehizadeh S, Hasanzad M, Kadijani AA, Akbari A (2019) The expression analysis of intestinal cancer stem cell marker Lgr5 in colorectal cancer patients and the correlation with histopathological markers. J Gastrointest Cancer. https://doi.org/10.1007/s12029-019-00295-w
Sato K, Uehara T, Iwaya M et al (2019) Correlation of clinicopathological features and LGR5 expression in colon adenocarcinoma. Ann Diagn Pathol 40:161–165. https://doi.org/10.1016/j.anndiagpath.2019.05.002
Shimokawa M, Ohta Y, Nishikori S et al (2017) Visualization and targeting of LGR5+ human colon cancer stem cells. Nature 545:187–192. https://doi.org/10.1038/nature22081
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69:7–34. https://doi.org/10.3322/caac.21551
Simon K (2016) Colorectal cancer development and advances in screening. Clin Interv Aging 11:967–976. https://doi.org/10.2147/CIA.S109285
Solomon H, Dinowitz N, Pateras IS et al (2018) Mutant p53 gain of function underlies high expression levels of colorectal cancer stem cells markers. Oncogene 37:1669–1684. https://doi.org/10.1038/s41388-017-0060-8
Sureban SM, May R, Mondalek FG et al (2011) Nanoparticle-based delivery of siDCAMKL-1 increases microRNA-144 and inhibits colorectal cancer tumor growth via a Notch-1 dependent mechanism. J Nanobiotechnol 9:40. https://doi.org/10.1186/1477-3155-9-40
Takahashi H, Ishii H, Nishida N et al (2011) Significance of Lgr5(+ve) cancer stem cells in the colon and rectum. Ann Surg Oncol 18:1166–1174. https://doi.org/10.1245/s10434-010-1373-9
Tao H, Tanaka T, Okabe K (2017) Doublecortin and CaM kinase-like-1 expression in pathological stage I non-small cell lung cancer. J Cancer Res Clin Oncol 143:1449–1459. https://doi.org/10.1007/s00432-017-2405-7
Uhlen M, Oksvold P, Fagerberg L et al (2010) Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28(12):1248–1250. https://doi.org/10.1038/nbt1210-1248
Walker F, Zhang H-H, Odorizzi A, Burgess AW (2011) LGR5 is a negative regulator of tumourigenicity, antagonizes Wnt signalling and regulates cell adhesion in colorectal cancer cell lines. PLoS ONE 6:e22733. https://doi.org/10.1371/journal.pone.0022733
Wang JL, Su WY, Lin YW et al (2017) CD44v6 overexpression related to metastasis and poor prognosis of colorectal cancer: a meta-analysis. Oncotarget. https://doi.org/10.18632/oncotarget.14163
Wang Q, Shi Y-L, Zhou K et al (2018a) PIK3CA mutations confer resistance to first-line chemotherapy in colorectal cancer. Cell Death Dis 9:739. https://doi.org/10.1038/s41419-018-0776-6
Wang X, Zhang X, Xu L et al (2018b) Expression of stem cell markers CD44 and Lgr5 in colorectal cancer and its relationship with lymph node and liver metastasis. Natl Med J China 98:2899–2904. https://doi.org/10.3760/cma.j.issn.0376-2491.2018.36.005
Wang Z, Tang Y, Xie L, et al (2019) The prognostic and clinical value of CD44 in colorectal cancer: a meta-analysis. Front Oncol
Whorton J, Sureban SM, May R et al (2015) DCLK1 is detectable in plasma of patients with Barrett’s esophagus and esophageal adenocarcinoma. Dig Dis Sci 60:509–513. https://doi.org/10.1007/s10620-014-3347-4
Wu W, Cao J, Ji Z et al (2016) Co-expression of Lgr5 and CXCR4 characterizes cancer stem-like cells of colorectal cancer. Oncotarget 7:81144–81155. https://doi.org/10.18632/oncotarget.13214
Wu X-S, Xi H-Q, Chen L (2012) Lgr5 is a potential marker of colorectal carcinoma stem cells that correlates with patient survival. World J Surg Oncol 10:244. https://doi.org/10.1186/1477-7819-10-244
Yang T, Peng H, Wang J et al (2013) Prognostic and diagnostic significance of annexin A2 in colorectal cancer. Colorectal Dis 15:e373–e381. https://doi.org/10.1111/codi.12207
Yeo W, Chan SL, Mo FKF et al (2015) Phase I/II study of temsirolimus for patients with unresectable Hepatocellular Carcinoma (HCC)—a correlative study to explore potential biomarkers for response. BMC Cancer 15:395. https://doi.org/10.1186/s12885-015-1334-6
Zarour LR, Anand S, Billingsley KG et al (2017) Colorectal cancer liver metastasis: evolving paradigms and future directions. Cmgh 3:163–173. https://doi.org/10.1016/j.jcmgh.2017.01.006
Zhang J, Cai H, Sun L et al (2018) LGR5, a novel functional glioma stem cell marker, promotes EMT by activating the Wnt/β-catenin pathway and predicts poor survival of glioma patients. J Exp Clin Cancer Res 37:225. https://doi.org/10.1186/s13046-018-0864-6
Zhang S, Chatterjee T, Godoy C et al (2019) GPR56 drives colorectal tumor growth and promotes drug resistance through upregulation of MDR1 expression via a RhoA-mediated mechanism. Mol Cancer Res. https://doi.org/10.1158/1541-7786.MCR-19-0436
Zheng Z, Yu H, Huang Q et al (2018) Heterogeneous expression of Lgr5 as a risk factor for focal invasion and distant metastasis of colorectal carcinoma. Oncotarget 9:30025–30033. https://doi.org/10.18632/oncotarget.23144
Zhou X, Geng L, Wang D et al (2017) R-Spondin1/LGR5 activates TGFβ signaling and suppresses colon cancer metastasis. Cancer Res 77:6589–6602. https://doi.org/10.1158/0008-5472.CAN-17-0219
Ziskin JL, Dunlap D, Yaylaoglu M et al (2013) In situ validation of an intestinal stem cell signature in colorectal cancer. Gut 62:1012–1023. https://doi.org/10.1136/gutjnl-2011-301195
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
AG, IZ, DJ, PA, and JD contributed to the experimental work, figures, and drafting of the manuscript. ŁS made substantial contributions to the design of the study and data analysis. JM, ED, and NS-M assisted with the experiments and data analysis. NA helped in drafting the manuscript as well as correction of the language. DG and ŁS supervised all the experiments. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
The study design was approved by the Local Bioethical Committee.
Informed consent
Informed consent was obtained from all patients and all clinical investigations were conducted according to the ethical and legal standards.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gzil, A., Zarębska, I., Jaworski, D. et al. The prognostic value of leucine-rich repeat-containing G-protein (Lgr5) and its impact on clinicopathological features of colorectal cancer. J Cancer Res Clin Oncol 146, 2547–2557 (2020). https://doi.org/10.1007/s00432-020-03314-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-020-03314-7