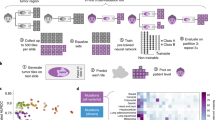
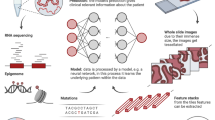
Abstract
Cancer of unknown primary (CUP) presents a complex diagnostic challenge, characterized by metastatic tumors of unknown tissue origin and a dismal prognosis. This review delves into the emerging significance of artificial intelligence (AI) and machine learning (ML) in transforming the landscape of CUP diagnosis, classification, and treatment. ML approaches, trained on extensive molecular profiling data, have shown promise in accurately predicting tissue of origin. Genomic profiling, encompassing driver mutations and copy number variations, plays a pivotal role in CUP diagnosis by providing insights into tumor type-specific oncogenic alterations. Mutational signatures (MS), reflecting somatic mutation patterns, offer further insights into CUP diagnosis. Known MS with established etiology, such as ultraviolet (UV) light-induced DNA damage and tobacco exposure, have been identified in cases of dedifferentiated/transdifferentiated melanoma and carcinoma. Deep learning models that integrate gene expression data and DNA methylation patterns offer insights into tissue lineage and tumor classification. In digital pathology, machine learning algorithms analyze whole-slide images to aid in CUP classification. Finally, precision oncology, guided by molecular profiling, offers targeted therapies independent of primary tissue identification. Clinical trials assigning CUP patients to molecularly guided therapies, including targetable alterations and tumor mutation burden as an immunotherapy biomarker, have resulted in improved overall survival in a subset of patients. In conclusion, AI- and ML-driven approaches are revolutionizing CUP management by enhancing diagnostic accuracy. Precision oncology utilizing enhanced molecular profiling facilitates the identification of targeted therapies that transcend the need to identify the tissue of origin, ultimately improving patient outcomes.


Similar content being viewed by others
Change history
22 December 2023
A Correction to this paper has been published: https://doi.org/10.1007/s00428-023-03728-x
References
Krämer A, Bochtler T, Pauli C et al (2023) Cancer of unknown primary: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up☆. Ann Oncol 34:228–246. https://doi.org/10.1016/j.annonc.2022.11.013
Pavlidis N, Pentheroudakis G (2012) Cancer of unknown primary site. The Lancet 379:1428–1435. https://doi.org/10.1016/S0140-6736(11)61178-1
Rassy E, Pavlidis N (2020) Progress in refining the clinical management of cancer of unknown primary in the molecular era. Nat Rev Clin Oncol 17:541–554. https://doi.org/10.1038/s41571-020-0359-1
Dermawan JK, Rubin BP (2021) The role of molecular profiling in the diagnosis and management of metastatic undifferentiated cancer of unknown primary✰: molecular profiling of metastatic cancer of unknown primary. Semin Diagn Pathol 38:193–198. https://doi.org/10.1053/j.semdp.2020.12.001
Tran KA, Kondrashova O, Bradley A et al (2021) Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med 13:152. https://doi.org/10.1186/s13073-021-00968-x
Shreve JT, Khanani SA, Haddad TC (2022) Artificial intelligence in oncology: current capabilities, future opportunities, and ethical considerations. Am Soc Clin Oncol Educ Book 842–851. https://doi.org/10.1200/EDBK_350652
Ciriello G, Miller ML, Aksoy BA et al (2013) Emerging landscape of oncogenic signatures across human cancers. Nat Genet 45:1127–1133. https://doi.org/10.1038/ng.2762
Ferreira I, Droop A, Edwards O et al (2021) The clinicopathologic spectrum and genomic landscape of de-/trans-differentiated melanoma. Mod Pathol 34:2009–2019. https://doi.org/10.1038/s41379-021-00857-z
Agaimy A, Stoehr R, Hornung A et al (2021) Dedifferentiated and undifferentiated melanomas: report of 35 new cases with literature review and proposal of diagnostic criteria. Am J Surg Pathol 45:240. https://doi.org/10.1097/PAS.0000000000001645
Chang JC, Alex D, Bott M et al (2019) Comprehensive next-generation sequencing unambiguously distinguishes separate primary lung carcinomas from intrapulmonary metastases: comparison with standard histopathologic approach. Clin Cancer Res 25:7113–7125. https://doi.org/10.1158/1078-0432.CCR-19-1700
Penson A, Camacho N, Zheng Y et al (2020) Development of genome-derived tumor type prediction to inform clinical cancer care. JAMA Oncol 6:84–91. https://doi.org/10.1001/jamaoncol.2019.3985
Sanjaya P, Maljanen K, Katainen R et al (2023) Mutation-Attention (MuAt): deep representation learning of somatic mutations for tumour typing and subtyping. Genome Med 15:47. https://doi.org/10.1186/s13073-023-01204-4
Posner A, Prall OW, Sivakumaran T et al (2023) A comparison of DNA sequencing and gene expression profiling to assist tissue of origin diagnosis in cancer of unknown primary. J Pathol 259:81–92. https://doi.org/10.1002/path.6022
Soh KP, Szczurek E, Sakoparnig T, Beerenwinkel N (2017) Predicting cancer type from tumour DNA signatures. Genome Med 9:104. https://doi.org/10.1186/s13073-017-0493-2
Nguyen L, Van Hoeck A, Cuppen E (2022) Machine learning-based tissue of origin classification for cancer of unknown primary diagnostics using genome-wide mutation features. Nat Commun 13:4013. https://doi.org/10.1038/s41467-022-31666-w
Jiao W, Atwal G, Polak P et al (2020) A deep learning system accurately classifies primary and metastatic cancers using passenger mutation patterns. Nat Commun 11:728. https://doi.org/10.1038/s41467-019-13825-8
Kandoth C, McLellan MD, Vandin F et al (2013) Mutational landscape and significance across 12 major cancer types. Nature 502:333–339. https://doi.org/10.1038/nature12634
Van Hoeck A, Tjoonk NH, van Boxtel R, Cuppen E (2019) Portrait of a cancer: mutational signature analyses for cancer diagnostics. BMC Cancer 19:457. https://doi.org/10.1186/s12885-019-5677-2
Hu X, Xu Z, De S (2020) Characteristics of mutational signatures of unknown etiology. NAR Cancer 2:zcaa026. https://doi.org/10.1093/narcan/zcaa026
Tothill RW, Li J, Mileshkin L et al (2013) Massively-parallel sequencing assists the diagnosis and guided treatment of cancers of unknown primary. J Pathol 231:413–423. https://doi.org/10.1002/path.4251
Varghese AM, Arora A, Capanu M et al (2017) Clinical and molecular characterization of patients with cancer of unknown primary in the modern era. Ann Oncol 28:3015–3021. https://doi.org/10.1093/annonc/mdx545
Mata DA, Williams EA, Sokol E et al (2022) Prevalence of UV mutational signatures among cutaneous primary tumors. JAMA Netw Open 5:e223833. https://doi.org/10.1001/jamanetworkopen.2022.3833
Kasago IS, Chatila WK, Lezcano CM et al (2023) Undifferentiated and dedifferentiated metastatic melanomas masquerading as soft tissue sarcomas: mutational signature analysis and immunotherapy response. Mod Pathol 36:100165. https://doi.org/10.1016/j.modpat.2023.100165
Bagge RO, Demir A, Karlsson J, et al (2018) Mutational signature and transcriptomic classification analyses as the decisive diagnostic tools for a cancer of unknown primary. JCO Precis Oncol 2:PO.18.00002. https://doi.org/10.1200/PO.18.00002
Wang Z, Zhang T, Wu W et al (2022) Detection and localization of solid tumors utilizing the cancer-type-specific mutational signatures. Front Bioeng Biotechnol 10:883791
Zhao Y, Pan Z, Namburi S et al (2020) CUP-AI-Dx: a tool for inferring cancer tissue of origin and molecular subtype using RNA gene-expression data and artificial intelligence. eBioMedicine 61:103030. https://doi.org/10.1016/j.ebiom.2020.103030
Hong J, Hachem LD, Fehlings MG (2022) A deep learning model to classify neoplastic state and tissue origin from transcriptomic data. Sci Rep 12:9669. https://doi.org/10.1038/s41598-022-13665-5
Grewal JK, Tessier-Cloutier B, Jones M et al (2019) Application of a neural network whole transcriptome–based pan-cancer method for diagnosis of primary and metastatic cancers. JAMA Netw Open 2:e192597. https://doi.org/10.1001/jamanetworkopen.2019.2597
Michuda J, Breschi A, Kapilivsky J et al (2023) Validation of a transcriptome-based assay for classifying cancers of unknown primary origin. Mol Diagn Ther 27:499–511. https://doi.org/10.1007/s40291-023-00650-5
Abraham J, Heimberger AB, Marshall J et al (2021) Machine learning analysis using 77,044 genomic and transcriptomic profiles to accurately predict tumor type. Transl Oncol 14:101016. https://doi.org/10.1016/j.tranon.2021.101016
Gardiner-Garden M, Frommer M (1987) CpG Islands in vertebrate genomes. J Mol Biol 196:261–282. https://doi.org/10.1016/0022-2836(87)90689-9
Sill M, Plass C, Pfister SM, Lipka DB (2020) Molecular tumor classification using DNA methylome analysis. Hum Mol Genet 29:R205–R213. https://doi.org/10.1093/hmg/ddaa147
Koelsche C, Schrimpf D, Stichel D et al (2021) Sarcoma classification by DNA methylation profiling. Nat Commun 12:498. https://doi.org/10.1038/s41467-020-20603-4
Capper D, Jones DTW, Sill M et al (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–474. https://doi.org/10.1038/nature26000
Giacopelli B, Wang M, Cleary A et al (2021) DNA methylation epitypes highlight underlying developmental and disease pathways in acute myeloid leukemia. Genome Res 31:747–761. https://doi.org/10.1101/gr.269233.120
Zhang Z, Lu Y, Vosoughi S et al (2023) HiTAIC: hierarchical tumor artificial intelligence classifier traces tissue of origin and tumor type in primary and metastasized tumors using DNA methylation. NAR Cancer 5:zcad017. https://doi.org/10.1093/narcan/zcad017
Zheng C, Xu R (2020) Predicting cancer origins with a DNA methylation-based deep neural network model. PLOS ONE 15:e0226461. https://doi.org/10.1371/journal.pone.0226461
Nagpal K, Foote D, Liu Y et al (2019) Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. Npj Digit Med 2:1–10. https://doi.org/10.1038/s41746-019-0112-2
Lu MY, Chen TY, Williamson DFK et al (2021) AI-based pathology predicts origins for cancers of unknown primary. Nature 594:106–110. https://doi.org/10.1038/s41586-021-03512-4
Saltz J, Gupta R, Hou L et al (2018) Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep 23:181-193.e7. https://doi.org/10.1016/j.celrep.2018.03.086
Ross JS, Sokol ES, Moch H et al (2021) Comprehensive genomic profiling of carcinoma of unknown primary origin: retrospective molecular classification considering the CUPISCO study design. The Oncologist 26:e394–e402. https://doi.org/10.1002/onco.13597
Pauli C, Bochtler T, Mileshkin L et al (2021) A challenging task: identifying patients with cancer of unknown primary (CUP) according to ESMO guidelines: the CUPISCO trial experience. The Oncologist 26:e769–e779. https://doi.org/10.1002/onco.13744
Hayashi H, Takiguchi Y, Minami H et al (2020) Site-specific and targeted therapy based on molecular profiling by next-generation sequencing for cancer of unknown primary site. JAMA Oncol 6:1–9. https://doi.org/10.1001/jamaoncol.2020.4643
Fusco MJ, Knepper TC, Balliu J et al (2022) Evaluation of targeted next-generation sequencing for the management of patients diagnosed with a cancer of unknown primary. The Oncologist 27:e9–e17. https://doi.org/10.1093/oncolo/oyab014
van Mourik A, Tonkin-Hill G, O’Farrell J et al (2023) Six-year experience of Australia’s first dedicated cancer of unknown primary clinic. Br J Cancer 129:301–308. https://doi.org/10.1038/s41416-023-02254-6
Cobain EF, Wu Y-M, Vats P et al (2021) Assessment of clinical benefit of integrative genomic profiling in advanced solid tumors. JAMA Oncol 7:525–533. https://doi.org/10.1001/jamaoncol.2020.7987
Fizazi K, Maillard A, Penel N et al (2019) A phase III trial of empiric chemotherapy with cisplatin and gemcitabine or systemic treatment tailored by molecular gene expression analysis in patients with carcinomas of an unknown primary (CUP) site (GEFCAPI 04). Ann Oncol 30:v851. https://doi.org/10.1093/annonc/mdz394
Marcus L, Fashoyin-Aje LA, Donoghue M et al (2021) FDA approval summary: pembrolizumab for the treatment of tumor mutational burden-high solid tumors. Clin Cancer Res Off J Am Assoc Cancer Res 27:4685–4689. https://doi.org/10.1158/1078-0432.CCR-21-0327
Chan TA, Yarchoan M, Jaffee E et al (2019) Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol Off J Eur Soc Med Oncol 30:44–56. https://doi.org/10.1093/annonc/mdy495
Marabelle A, Fakih M, Lopez J et al (2020) Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol 21:1353–1365. https://doi.org/10.1016/S1470-2045(20)30445-9
Sha D, Jin Z, Budzcies J et al (2020) Tumor mutational burden (TMB) as a predictive biomarker in solid tumors. Cancer Discov 10:1808–1825. https://doi.org/10.1158/2159-8290.CD-20-0522
Buchhalter I, Rempel E, Endris V et al (2019) Size matters: dissecting key parameters for panel-based tumor mutational burden analysis. Int J Cancer 144:848–858. https://doi.org/10.1002/ijc.31878
Author information
Authors and Affiliations
Contributions
SWL wrote the first draft of the manuscript. JKD conceived the project, collected references, wrote, and edited the manuscript. BPR oversaw the project, wrote, and edited the manuscript. All authors approve of the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The first name of the first author has been corrected.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lorkowski, S.W., Dermawan, J.K. & Rubin, B.P. The practical utility of AI-assisted molecular profiling in the diagnosis and management of cancer of unknown primary: an updated review. Virchows Arch 484, 369–375 (2024). https://doi.org/10.1007/s00428-023-03708-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-023-03708-1