Abstract
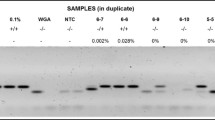
The present study was undertaken to explore and validate novel hypermethylated DNA regions in squamous cell carcinoma of the tongue (SCCT). Genome-wide methylation changes were identified by differential methylation hybridization (DMH) microarray and validated by bisulfite genome sequencing (BGS). The results were compared against datasets from The Cancer Genome Atlas head and neck squamous cell carcinoma (TCGA-HNSCC), Gene Expression Omnibus (GSE26549), and ArrayExpress (E-MTAB-1328). DMH identified 116 hypomethylated and 241 hypermethylated regions. Of the latter, 24 were localized to promoter or 5′-UTR regions. By BGS, promoter sequences of DAPK1, LRPPRC, RAB6C, and ZNF471 were significantly hypermethylated in tumors when compared with matched normal tissues (P < 0.0001). A TCGA-HNSCC dataset (516 cases of cancer and 50 normal tissue samples) further confirmed hypermethylation of DAPK1, RAB6C, and ZNF471. Sensitivity and specificity of methylation markers for a diagnosis of cancer were in the range of 70–100% in our study and from TCGA-HNSCC datasets, with an area under curve (AUC) of 0.83 and above. Kaplan-Meier survival analysis of TCGA-HNSCC expression data revealed that patients with low expressions of DAPK1, RAB6C, and ZNF471 showed poorer survival than patients with high expression (P = 0.02). Human papillomavirus (HPV) was found in 55% of cases, HPV16 being the predominant genotype. DAPK1 immunohistochemical staining was lower in SCCT than in normal buccal epithelial cells. This is the first study to report hypermethylation of LRPPRC, RAB6C, and ZNF471 in SCCT and its diagnostic and prognostic potentials in a specific head and neck squamous cell carcinoma.




Similar content being viewed by others
References
Daley T, Darling M (2003) Nonsquamous cell malignant tumours of the oral cavity: an overview. J Can Dent Assoc 69(9):577–582
Parkin DM, Pisani P, Ferlay J (1999) Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer 80(6):827–841
Gupta S, Singh R, Gupta OP, Tripathi A (2014) Prevalence of oral cancer and pre-cancerous lesions and the association with numerous risk factors in North India: a hospital based study. Natl J Maxillofac Surg 5(2):142–148. doi:10.4103/0975-5950.154816
Mishra A, Meherotra R (2014) Head and neck cancer: global burden and regional trends in India. Asian Pacific Journal of Cancer Prevention : APJCP 15(2):537–550
Kuriakose M, Sankaranarayanan M, Nair MK, Cherian T, Sugar AW, Scully C, Prime SS (1992) Comparison of oral squamous cell carcinoma in younger and older patients in India. Eur J Cancer B Oral Oncol 28B(2):113–120
Llewellyn CD, Johnson NW, Warnakulasuriya KA (2001) Risk factors for squamous cell carcinoma of the oral cavity in young people—a comprehensive literature review. Oral Oncol 37(5):401–418
Pathak KA, Das AK, Agarwal R, Talole S, Deshpande MS, Chaturvedi P, Pai PS, Chaukar DA, D’Cruz AK (2006) Selective neck dissection (I–III) for node negative and node positive necks. Oral Oncol 42(8):837–841. doi:10.1016/j.oraloncology.2005.12.002
Ram H, Sarkar J, Kumar H, Konwar R, Bhatt ML, Mohammad S (2011) Oral cancer: risk factors and molecular pathogenesis. J Maxillofac Oral Surg 10(2):132–137. doi:10.1007/s12663-011-0195-z
Krishnan NM, Dhas K, Nair J, Palve V, Bagwan J, Siddappa G, Suresh A, Kekatpure VD, Kuriakose MA, Panda B (2016) A minimal DNA methylation signature in oral tongue squamous cell carcinoma links altered methylation with tumor attributes. Mol Cancer Res. doi:10.1158/1541-7786.MCR-15-0395
Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Trevino L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN (2011) Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333(6046):1154–1157. doi:10.1126/science.1206923
Radhakrishnan R, Kabekkodu S, Satyamoorthy K (2011) DNA hypermethylation as an epigenetic mark for oral cancer diagnosis. J Oral Pathol Med 40(9):665–676. doi:10.1111/j.1600-0714.2011.01055.x
Hu Z, Zhong Z, Huang S, Wen H, Chen X, Chu H, Li Q, Sun C (2016) Decreased expression of Beclin1 is significantly associated with a poor prognosis in oral tongue squamous cell carcinoma. Mol Med Rep. doi:10.3892/mmr.2016.5437
Peng CH, Liao CT, Peng SC, Chen YJ, Cheng AJ, Juang JL, Tsai CY, Chen TC, Chuang YJ, Tang CY, Hsieh WP, Yen TC (2011) A novel molecular signature identified by systems genetics approach predicts prognosis in oral squamous cell carcinoma. PLoS One 6(8):e23452. doi:10.1371/journal.pone.0023452
Agodi A, Barchitta M, Quattrocchi A, Maugeri A, Vinciguerra M (2015) DAPK1 promoter methylation and cervical cancer risk: a systematic review and a meta-analysis. PLoS One 10(8):e0135078. doi:10.1371/journal.pone.0135078
Chen HC, Yang CM, Cheng JT, Tsai KW, Fu TY, Liou HH, Tseng HH, Lee JH, Li GC, Wang JS, Hou YY, Weng TJ, Ger LP (2016) Global DNA hypomethylation is associated with the development and poor prognosis of tongue squamous cell carcinoma. J Oral Pathol Med 45(6):409–417. doi:10.1111/jop.12381
Nagata S, Hamada T, Yamada N, Yokoyama S, Kitamoto S, Kanmura Y, Nomura M, Kamikawa Y, Yonezawa S, Sugihara K (2012) Aberrant DNA methylation of tumor-related genes in oral rinse: a noninvasive method for detection of oral squamous cell carcinoma. Cancer 118(17):4298–4308. doi:10.1002/cncr.27417
Ovchinnikov DA, Cooper MA, Pandit P, Coman WB, Cooper-White JJ, Keith P, Wolvetang EJ, Slowey PD, Punyadeera C (2012) Tumor-suppressor gene promoter hypermethylation in saliva of head and neck cancer patients. Transl Oncol 5(5):321–326
Rettori MM, de Carvalho AC, Bomfim Longo AL, de Oliveira CZ, Kowalski LP, Carvalho AL, Vettore AL (2013) Prognostic significance of TIMP3 hypermethylation in post-treatment salivary rinse from head and neck squamous cell carcinoma patients. Carcinogenesis 34(1):20–27. doi:10.1093/carcin/bgs311
Zhang S, Feng XL, Shi L, Gong CJ, He ZJ, Wu HJ, Ling TY (2013) Genome-wide analysis of DNA methylation in tongue squamous cell carcinoma. Oncol Rep 29(5):1819–1826. doi:10.3892/or.2013.2309
Jithesh PV, Risk JM, Schache AG, Dhanda J, Lane B, Liloglou T, Shaw RJ (2013) The epigenetic landscape of oral squamous cell carcinoma. Br J Cancer 108(2):370–379. doi:10.1038/bjc.2012.568
Huang TH, Perry MR, Laux DE (1999) Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet 8(3):459–470
Kabekkodu SP, Bhat S, Radhakrishnan R, Aithal A, Mascarenhas R, Pandey D, Rai L, Kushtagi P, Mundyat GP, Satyamoorthy K (2014) DNA promoter methylation-dependent transcription of the double C2-like domain beta (DOC2B) gene regulates tumor growth in human cervical cancer. J Biol Chem 289(15):10637–10649. doi:10.1074/jbc.M113.491506
Rotti H, Mallya S, Kabekkodu SP, Chakrabarty S, Bhale S, Bharadwaj R, Bhat BK, Dedge AP, Dhumal VR, Gangadharan GG, Gopinath PM, Govindaraj P, Joshi KS, Kondaiah P, Nair S, Nair SN, Nayak J, Prasanna BV, Shintre P, Sule M, Thangaraj K, Patwardhan B, Valiathan MV, Satyamoorthy K (2015) DNA methylation analysis of phenotype specific stratified Indian population. J Transl Med 13:151. doi:10.1186/s12967-015-0506-0
Fuessel Haws AL, He Q, Rady PL, Zhang L, Grady J, Hughes TK, Stisser K, Konig R, Tyring SK (2004) Nested PCR with the PGMY09/11 and GP5(+)/6(+) primer sets improves detection of HPV DNA in cervical samples. J Virol Methods 122(1):87–93. doi:10.1016/j.jviromet.2004.08.007
Kabekkodu SP, Bhat S, Pandey D, Varghese VK, Shukla V, Ghosh S, Kushtagi P, Bhat P, Gopinath PM, Satyamoorthy K (2015) Prevalence of human papillomavirus types and phylogenetic analysis of HPV-16 L1 variants from southern India. Asian Pacific journal of cancer prevention : APJCP 16(5):2073–2080
Calmon MF, Colombo J, Carvalho F, Souza FP, Filho JF, Fukuyama EE, Camargo AA, Caballero OL, Tajara EH, Cordeiro JA, Rahal P (2007) Methylation profile of genes CDKN2A (p14 and p16), DAPK1, CDH1, and ADAM23 in head and neck cancer. Cancer Genet Cytogenet 173(1):31–37. doi:10.1016/j.cancergencyto.2006.09.008
Missaoui N, Hmissa S, Trabelsi A, Traore C, Mokni M, Dante R, Frappart L (2011) Promoter hypermethylation of CDH13, DAPK1 and TWIST1 genes in precancerous and cancerous lesions of the uterine cervix. Pathol Res Pract 207(1):37–42. doi:10.1016/j.prp.2010.11.001
Li X, Lv L, Zheng J, Zhou J, Liu B, Chen H, Liang C, Wang R, Su L, Li X, Fan D (2014) The significance of LRPPRC overexpression in gastric cancer. Med Oncol 31(2):818. doi:10.1007/s12032-013-0818-y
Sato N, Koinuma J, Ito T, Tsuchiya E, Kondo S, Nakamura Y, Daigo Y (2010) Activation of an oncogenic TBC1D7 (TBC1 domain family, member 7) protein in pulmonary carcinogenesis. Genes Chromosomes Cancer 49(4):353–367. doi:10.1002/gcc.20747
Takahashi F, Chiba N, Tajima K, Hayashida T, Shimada T, Takahashi M, Moriyama H, Brachtel E, Edelman EJ, Ramaswamy S, Maheswaran S (2011) Breast tumor progression induced by loss of BTG2 expression is inhibited by targeted therapy with the ErbB/HER inhibitor lapatinib. Oncogene 30(27):3084–3095. doi:10.1038/onc.2011.24
Serenaite I, Daniunaite K, Jankevicius F, Laurinavicius A, Petroska D, Lazutka JR, Jarmalaite S (2014) Heterogeneity of DNA methylation in multifocal prostate cancer. Virchows Arch 466(1):53–59. doi:10.1007/s00428-014-1678-3
Toffolatti L, Scquizzato E, Cavallin S, Canal F, Scarpa M, Stefani PM, Gherlinzoni F, Dei Tos AP (2014) MGMT promoter methylation and correlation with protein expression in primary central nervous system lymphoma. Virchows Arch 465(5):579–586. doi:10.1007/s00428-014-1622-6
Rhee YY, Lee TH, Song YS, Wen X, Kim H, Jheon S, Lee CT, Kim J, Cho NY, Chung JH, Kang GH (2015) Prognostic significance of promoter CpG island hypermethylation and repetitive DNA hypomethylation in stage I lung adenocarcinoma. Virchows Arch 466(6):675–683. doi:10.1007/s00428-015-1749-0
Martinez-Glez V, Franco-Hernandez C, Gonzalez-Gomez P, Isla A, De Campos JM, Vaquero J, Gutierrez M, Casartelli C, Rey JA (2007) DAPK1 promoter hypermethylation in brain metastases and peripheral blood. Neoplasma 54(2):123–126
Supic G, Kozomara R, Jovic N, Zeljic K, Magic Z (2011) Prognostic significance of tumor-related genes hypermethylation detected in cancer-free surgical margins of oral squamous cell carcinomas. Oral Oncol 47(8):702–708. doi:10.1016/j.oraloncology.2011.05.014
Marttila S, Kananen L, Hayrynen S, Jylhava J, Nevalainen T, Hervonen A, Jylha M, Nykter M, Hurme M (2015) Ageing-associated changes in the human DNA methylome: genomic locations and effects on gene expression. BMC Genomics 16:179. doi:10.1186/s12864-015-1381-z
Mitchell SM, Ross JP, Drew HR, Ho T, Brown GS, Saunders NF, Duesing KR, Buckley MJ, Dunne R, Beetson I, Rand KN, McEvoy A, Thomas ML, Baker RT, Wattchow DA, Young GP, Lockett TJ, Pedersen SK, Lapointe LC, Molloy PL (2014) A panel of genes methylated with high frequency in colorectal cancer. BMC Cancer 14:54. doi:10.1186/1471-2407-14-54
Xu F, Morin C, Mitchell G, Ackerley C, Robinson BH (2004) The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome C oxidase) I and COX III mRNA. Biochem J 382(Pt 1):331–336. doi:10.1042/BJ20040469
Cooper MP, Qu L, Rohas LM, Lin J, Yang W, Erdjument-Bromage H, Tempst P, Spiegelman BM (2006) Defects in energy homeostasis in Leigh syndrome French Canadian variant through PGC-1alpha/LRP130 complex. Genes Dev 20(21):2996–3009. doi:10.1101/gad.1483906
Cooper MP, Uldry M, Kajimura S, Arany Z, Spiegelman BM (2008) Modulation of PGC-1 coactivator pathways in brown fat differentiation through LRP130. J Biol Chem 283(46):31960–31967. doi:10.1074/jbc.M805431200
Tsuchiya N, Fukuda H, Nakashima K, Nagao M, Sugimura T, Nakagama H (2004) LRP130, a single-stranded DNA/RNA-binding protein, localizes at the outer nuclear and endoplasmic reticulum membrane, and interacts with mRNA in vivo. Biochem Biophys Res Commun 317(3):736–743. doi:10.1016/j.bbrc.2004.03.103
Tian T, Ikeda J, Wang Y, Mamat S, Luo W, Aozasa K, Morii E (2012) Role of leucine-rich pentatricopeptide repeat motif-containing protein (LRPPRC) for anti-apoptosis and tumourigenesis in cancers. Eur J Cancer 48(15):2462–2473. doi:10.1016/j.ejca.2012.01.018
Zhen Y, Stenmark H (2015) Cellular functions of Rab GTPases at a glance. J Cell Sci 128(17):3171–3176. doi:10.1242/jcs.166074
Shan J, Yuan L, Budman DR, Xu HP (2002) WTH3, a new member of the Rab6 gene family, and multidrug resistance. Biochim Biophys Acta 1589(2):112–123
Tian K, Jurukovski V, Yuan L, Shan J, Xu H (2005) WTH3, which encodes a small G protein, is differentially regulated in multidrug-resistant and sensitive MCF7 cells. Cancer Res 65(16):7421–7428. doi:10.1158/0008-5472.CAN-05-0658
Abogunrin S, Di Tanna GL, Keeping S, Carroll S, Iheanacho I (2014) Prevalence of human papillomavirus in head and neck cancers in European populations: a meta-analysis. BMC Cancer 14:968. doi:10.1186/1471-2407-14-968
Chaitanya NC, Allam NS, Gandhi Babu DB, Waghray S, Badam RK, Lavanya R (2016) Systematic meta-analysis on association of human papilloma virus and oral cancer. J Cancer Res Ther 12(2):969–974. doi:10.4103/0973-1482.179098
Morbini P, Benazzo M (2016) Human papillomavirus and head and neck carcinomas: focus on evidence in the babel of published data. Acta otorhinolaryngologica Italica : organo ufficiale della Societa italiana di otorinolaringologia e chirurgia cervico-facciale 36(4):249–258. doi:10.14639/0392-100X-853
Acknowledgements
Support for this work by Indian Council of Medical Research (ICMR), Government of India; FIST, Department of Science and Technology (DST), Government of India; Vision Group on Science and Technology (K-FIST level II), BTFS Program, Government of Karnataka; and Manipal University are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The study protocol was approved by the institutional ethics committee of Manipal University and was conducted in compliance with The Code of Ethics of the World Medical Association (Declaration of Helsinki). Samples were collected after obtaining the written consent document for participation in the study.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(DOCX 16 kb)
ESM 2
(DOCX 20 kb)
Supplementary Fig. 1
Methylation and expression analysis on TCGA dataset. A) The methylation analysis was performed on 516 tumor and 50 normal samples. The frequency of hypermethylation was significantly higher for DAPK1, RAB6C and ZNF471 in tumor samples as opposed to normal samples by Fisher’s exact test. P ≤ 0.05 was considered as statistically significant. Sample with β value above 0.2 was considered as methylated. B) Methylation analysis in matched normal and tumor samples (50 samples each). C) The line graph shows methylation of individual CpG sites. Except LRPPRC, all three genes showed significant hypermethylation in specific CpG sites. D) Expression analysis of normal and tumor samples from TCGA HNSCC dataset. Both methylation and expression analysis were performed using Wanderer tool. Statistical analysis was performed by Fisher’s exact test using GraphPad Prism software. (JPEG 476 kb)
Supplementary Fig. 2
The hierarchical clustering and PCA analysis. The clustering and PCA analysis was performed using Partek Genomic suite software. Hierarchical clustering was performed by Euclidean distance and average linkage method. A) Hierarchical clustering of DAPK1, LRPPRC, RAB6C and ZNF471 in all the samples (left) and in matched normal and tumor samples (right) showing clear discrimination power of DNA methylation to distinguish between normal and tumor samples B) PCA analysis of all samples (left) and in matched samples (right) respectively. The use of matched samples for clustering increased mapping from 86.8% to 90.8%. C) Hierarchical clustering using DAPK1, RAB6C and ZNF471 in all samples (left) and matched samples (right). D) The corresponding PCA in all the samples (left) and matched samples (right). PCA analysis using three gene combinations increased PCA mapping to 100% in both matched and non-matched samples respectively. (JPEG 616 kb)
Supplementary Fig. 3
Survival analysis in patients head and neck cancer. The samples were classified into high expressing and low expressing DAPK1, LRPPRC, RAB6C and ZNF471 cases. Survival analysis was performed by SurvExpress software using TCGA-HNSCC, GSE26549 and E-MTAB-1328 datasets. Censoring samples are shown as “+” marks. Horizontal axis represents time to event. Dataset, outcome event, time scale, concordance index (CI), and p-value of the log-rank test are shown. Red and Green curves denote high- and low-expressing groups (indicated in the graphs as “high” and “low” risks) respectively. The number of individuals, the number of censored, and the CI of each group are shown in the top-right insets. A) Represents Kaplan-Meier curve for expressing groups, concordance index, and p-value of the log-rank testing equality of survival curves from TCGA-HNSCC. In G3 and T2 patients of TCGA-HNSCC had poor survival in high expressing category as opposed to low expressing category (P ≤ 0.005). B) Represents Kaplan Meier survival analysis using GSE26549 dataset. High expressing groups showed significantly lower survival as opposed to low expressing group. Additionally p63 high expression group had significantly lower survival. Moreover, in the high hypermethylated gene expressing group, those with high p63 expression had significantly poorer survival than those with low p63 expression. (JPEG 299 kb)
Rights and permissions
About this article
Cite this article
Bhat, S., Kabekkodu, S.P., Jayaprakash, C. et al. Gene promoter-associated CpG island hypermethylation in squamous cell carcinoma of the tongue. Virchows Arch 470, 445–454 (2017). https://doi.org/10.1007/s00428-017-2094-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-017-2094-2