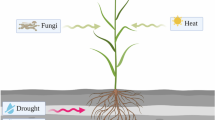
Abstract
Main conclusion
This review provides a comprehensive overview of the CRISPR/Cas9 technique and the research areas of this gene editing tool in improving wheat quality.
Abstract
Wheat (Triticum aestivum L.), the basic nutrition for most of the human population, contributes 20% of the daily energy needed because of its, carbohydrate, essential amino acids, minerals, protein, and vitamin content. Wheat varieties that produce high yields and have enhanced nutritional quality will be required to fulfill future demands. Hexaploid wheat has A, B, and D genomes and includes three like but not identical copies of genes that influence important yield and quality. CRISPR/Cas9, which allows multiplex genome editing provides major opportunities in genome editing studies of plants, especially complicated genomes such as wheat. In this overview, we discuss the CRISPR/Cas9 technique, which is credited with bringing about a paradigm shift in genome editing studies. We also provide a summary of recent research utilizing CRISPR/Cas9 to investigate yield, quality, resistance to biotic/abiotic stress, and hybrid seed production. In addition, we provide a synopsis of the laboratory experience-based solution alternatives as well as the potential obstacles for wheat CRISPR studies. Although wheat’s extensive genome and complicated polyploid structure previously slowed wheat genetic engineering and breeding progress, effective CRISPR/Cas9 systems are now successfully used to boost wheat development.

Similar content being viewed by others
Data availability
Because no data sets were generated or analyzed during the current study, data sharing does not apply to this article.
References
Aadel H, Udupa S, Abdelwahd R, Gaboun F, Diria G, Douira A, Iraqi D (2021) Agrobacterium-mediated genetic transformation of bread wheat (Triticum aestivum L.) using immature embryos. Rom Agric Res 38:99–107
Abdallah NA, Elsharawy H, Abulela HA, Thilmony R, Abdelhadi AA (2022) Elarabi NI (2022) Multiplex CRISPR/Cas9-mediated genome editing to address drought tolerance in wheat. GM Crops Food. https://doi.org/10.1080/216456982120313
Abdul R, Ishfaq AH, Imran M, Azhar H (2011) Development of in planta transformation protocol for wheat. Afr J Biotechnol 10:740–750
Abe F, Haque E, Hisano H, Tanaka T, Kamiya Y, Mikami M, Kawaura K, Endo M, Onishi K, Hayashi T, Sato K (2019) Genome-edited triple-recessive mutation alters seed dormancy in wheat. Cell Rep 28:1362–1369
Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semanova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F (2016) C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Sci. https://doi.org/10.1126/science.aaf5573
Achary V, Reddy MK (2021) CRISPR-Cas9 mediated mutation in GRAIN WIDTH and WEIGHT2 (GW2) locus improves rice’s aleurone layer and grain nutritional quality. Sci Rep 11:1–13
Aglawe SB, Barbadikar KM, Mangrauthia SK, Madhav MS (2018) New breeding technique “genome editing” for crop improvement: applications, potentials and challenges. 3 Biotech 8:1–20
Ahansal K, Abdelwahd R, Udupa S, Aadel H, Gaboun F, Ibriz M, Iraqi D (2022) Effect of type of mature embryo explants and acetosyringone on Agrobacterium-mediated transformation of moroccan durum wheat. J Biosci 38:e38007
Ahmad HM, Iqbal MS, Abdullah M, El-Tabakh MA, Oranab S, Mudassar M, Shimira F, Zahid G (2023) Recent Trends in Genome Editing Technologies for Agricultural Crop Improvement. Sustainable Agriculture in the Era of the OMICs Revolution. Springer International Publishing, Cham, pp 357–379
Alikina O, Chernobrovkina M, Dolgov S, Miroshnichenko D (2016) Tissue culture efficiency of wheat species with different genomic formulas. Crop Breed Appl Biotechnol 16:307–314
Allahi S, Khodaparast SA, Sohani MM (2014) Agrobacterium-mediated transformation of Indica rice: A non-tissue culture approach. Int J Agric Innov Res 3:2319–1473
Amoah BK, Wu H, Sparks C, Jones HD (2001) Factors influencing Agrobacterium-mediated transient expression of uid A in wheat inflorescence tissue. J Exp Bot 52:1135–1142
Anas M, Liao F, Verma KK, Sarwar MA, Mahmood A, Chen ZL, Li Q, Zeng XP, Li YR (2020) Fate of nitrogen in agriculture and environment: agronomic, eco-physiological and molecular approaches to improve nitrogen use efficiency. Biol Res 53:1–20
Anders C, Niewoehner O, Duerst A, Jinek M (2014) Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513:569–573
Appels R, Eversole K, Feuille C, Keller B, Rogers J, Stein N (2018) The International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:10–1126
Arndell T, Sharma N, Langridge P, Baumann U, Watson-Haigh NS, Whitford R (2019) gRNA validation for wheat genome editing with the CRISPR-Cas9 system. BMC Biotechnol 19:1–12
Arslan E, Agar G, Aydin M (2021) Humic acid as a biostimulant in improving drought tolerance in wheat: The expression patterns of drought-related genes. Plant Mol Biol Rep 39:508–519
Awan MJA, Pervaiz K, Rasheed A, Amin I, Saeed NA, Dhugga KS, Mansoor S (2022) Genome edited wheat-current advances for the second green revolution. Biotechnol Adv 60:108006
Aydin M, Tosun M, Haliloglu K (2011) Plant regeneration in wheat mature embryo culture. Afr J Biotechnol 10:15749–15755
Aydin M, Pour AH, Haliloğlu K, Tosun M (2016) Effect of polyamines on somatic embryogenesis via mature embryo in wheat. Turk J Biol 40:1178–1184
Aydin M, Arslan E, Yigider E, Taspinar MS, Agar G (2021) Protection of Phaseolus vulgaris L. from Herbicide 2, 4-D results from exposing seeds to humic acid. Arab J Sci Eng 46:163–173
Badhan S, Ball AS, Mantri N (2021) First report of CRISPR/Cas9 mediated DNA-free editing of 4CL and RVE7 genes in chickpea protoplasts. Int J Mol Sci 22:396
Bae S, Park J, Kim JS (2014) Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30:1473–1475
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712
Baskar G, Aiswarya R (2018) Overview on mitigation of acrylamide in starchy fried and baked foods. J Sci Food Agric 98:4385–4394
Bettgenhaeuser J, Krattinger SG (2019) Rapid gene cloning in cereals. Theor Appl Genet 132:699–711
Blin K, Pedersen LE, Weber T, Lee SY (2016) CRISPy-web: an online resource to design sgRNAs for CRISPR applications. Synth Syst Biotechnol 1:118–121
Borrelli VM, Brambilla V, Rogowsky P, Marocco A, Lanubile A (2018) The enhancement of plant disease resistance using CRISPR/Cas9 technology. Front Plant Sci 9:1245
Brauer EK, Balcerzak M, Rocheleau H, Leung W, Schernthaner J, Subramaniam R, Ouellet T (2020) Genome editing of a deoxynivalenol-induced transcription factor confers resistance to Fusarium graminearum in wheat. Mol Plant Microbe Interact 33:553–560
Caruso SM, Quinn PM, da Costa BL, Tsang SH (2022) CRISPR/Cas therapeutic strategies for autosomal dominant disorders. Clin Drug Investig 132:e158287
Carvalho CHS, Zehr UB, Gunaratna N, Anderson J, Kononowicz HH, Hodges TK, Axtell JD (2004) Agrobacterium-mediated transformation of sorghum: factors that affect transformation efficiency. Genet Mol Biol 27:259–269
Chang H, Yi B, Ma R, Zhang X, Zhao H, Xi Y (2016) CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci Rep 6:1–12
Chanyalew S, Assefa K, Tadele Z (2019) Tef [Eragrostis tef (Zucc.) Trotter] Breeding. Adv Plant Breed Strateg Cereals 5:373–403
Chaudhary J, Alisha A, Bhatt V, Chandanshive S, Kumar N, Mir Z, Kumar A, Yadav SK, Shivaraj SM, Sonah H, Deshmukh R (2019) Mutation breeding in tomato: Advances, applicability and challenges. Plants 8:128
Chauhan H, Khurana P (2017) Wheat genetic transformation using mature embryos as explants. In Wheat Biotechnology. Humana Press, New York pp. 153–167
Chavez M, Chen X, Finn PB, Qi LS (2022) Advances in CRISPR therapeutics. Nat Rev Nephrol 1–14
Chen K, Wang Y, Zhang R, Zhang H, Gao C (2019a) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70:667–697
Chen W, Zhang H, Zhang Y, Wang Y, Gan J, Ji Q (2019b) Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease. PLoS Biol 17:e3000496
Chen Z, Ke W, He F, Chai L, Cheng X, Xu H, Wang X, Du D, Zhao Y, Chen X, Xing J, Xin M, Guo W, Hu Z, Su Z, Liu J, Peng H, Yao Y, Sun Q, Ni Z (2022) A single nucleotide deletion in the third exon of FT-D1 increases the spikelet number and delays heading date in wheat (Triticum aestivum L.). Plant Biotechnol J 20:920
Chen Z, Debernardi JM, Dubcovsky J, Gallavotti A (2022a) Recent advances in crop transformation technologies. Nat Plants 1–9
Cheng M, Fry JE, Pang S, Zhou H, Hironaka CM, Duncan DR, Conner TW, Wan Y (1997) Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol 115:971–980
Cheng X, Gao C, Liu X, Xu D, Pan X, Gao W, Yan S, Yao H, Cao J, Min X, Lu J, Chang C, Zhang H, Ma C (2022) Characterization of the wheat VQ protein family and expression of candidate genes associated with seed dormancy and germination. BMC Plant Biol 22:1–18
Chylinski K, Le Rhun A, Charpentier E (2013) The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol 10:726–737
Cram D, Kulkarni M, Buchwaldt M, Rajagopalan N, Bhowmik P, Rozwadowski K, Parkin İAP, Sharpe AG, Kagale S (2019) WheatCRISPR: a web-based guide RNA design tool for CRISPR/Cas9-mediated genome editing in wheat. BMC Plant Biol 19:1–8
Curtis BC, Rajaram S, Gómez Macpherson H (2002) Bread wheat: improvement and production. Food and Agriculture Organization of the United Nations (FAO)
Dai SH, Zheng P, Marmey P, Zhang SP, Tian WZ, Chen SY, Beachy RN, Fauquet C (2001) Comparative analysis of transgenic rice plants obtained by Agrobacterium-mediated transformation and particle bombardment. Mol Breed 7:25–33
Dan Y (2008) Biological functions of antioxidants in plant transformation. In Vitro Cell Dev Biol 44:149–161
Das D, Reddy M, Upadhyaya K, Sopory S (2002) An efficient leaf-disc culture method for the regeneration via somatic embryogenesis and transformation of grape (Vitis vinifera L.). Plant Cell Rep 20:999–1005
Dayani S, Sabzalian MR, Mazaheri-Tirani M (2019) CRISPR/Cas9 genome editing in bread wheat (Triticum aestivum L.) genetic improvement. Adv Plant Breed Strateg Cereals 5:453–469
De Saeger J, Park J, Chung HS, Hernalsteens JP, Van Lijsebettens M, Inzé D, Montagu MV, Depuydt S (2021) Agrobacterium strains and strain improvement: Present and outlook. Biotechnol Adv 53:107677
Ding L, Li S, Gao J, Wang Y, Yang G, He G (2009) Optimization of Agrobacterium-mediated transformation conditions in mature embryos of elite wheat. Mol Biol Rep 36:29–36
Ding Y, Li H, Chen LL, Xie K (2016) Recent advances in genome editing using CRISPR/Cas9. Front Plant Sci 7:703
Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096
Erturk FA, Agar G, Arslan E, Nardemir G (2015) Analysis of genetic and epigenetic effects of maize seeds in response to heavy metal (Zn) stress. Environ Sci Pollut Res 22:10291–10297
Esmail SM, Omar GE, Mourad AM (2023) In-Depth Understanding of the Genetic Control of Stripe Rust Resistance (Puccinia striiformis f. sp. tritici) Induced in Wheat (Triticum aestivum) by Trichoderma asperellum T34. Plant Dis 107:457–472
FAO (2022) The state of food security and nutrition in the world 2020: transforming food systems for affordable healthy diets. Food & Agriculture Org. 2022. Available from: https://www.fao.org/3/ca9692en/online/ca9692en.html
Fedorova I, Vasileva A, Selkova P, Abramova M, Arseniev A, Pobegalov G, Kazalov M, Musharova O, Goryanin I, Artamonova D, Zyubko T, Shmakov S, Artamonova T, Khodorkovskii M, Severinov K (2020) PpCas9 from Pasteurella pneumotropica—a compact Type II-C Cas9 ortholog active in human cells. Nucleic Acids Res 48:12297–12309
Gao XQ, Wang N, Wang XL, Zhang XS (2019) Architecture of wheat inflorescence: insights from rice. Trends Plant Sci 24:802–809
Garneau JE, Dupuis M-E, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71
Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. PNAS 109:E2579–E2586
Gil-Humanes J, Wang Y, Liang Z, Shan Q, Ozuna CV, Sánchez-León S, Baltes NJ, Starker C, Barro F, Gao C, Voytas DF (2017) High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J 89:1251–1262
González Castro N, Bjelic J, Malhotra G, Huang C, Alsaffar SH (2021) Comparison of the feasibility, efficiency, and safety of genome editing technologies. Int J Mol Sci 22:10355
Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F (2018) Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360:439–444
Gostimskaya I (2022) CRISPR–Cas9: A History of Its Discovery and Ethical Considerations of Its Use in Genome Editing. Biochem (Moscow) 87:777–788
Greer MS, Kovalchuk I, Eudes F (2009) Ammonium nitrate improves direct somatic embryogenesis and biolistic transformation of Triticum aestivum. N Biotechnol 26:44–52
Guha TK, Edgell DR (2017) Applications of alternative nucleases in the age of CRISPR/Cas9. Int J Mol Sci 18:2565
Gupta D, Bhattacharjee O, Mandal D, Sen MK, Dey D, Dasgupta A, Kazi TA, Gupta R, Sinharoy S, Acharya K, Chattopadhyay D, Ravichandiran V, Roy S, Ghosh D (2019) CRISPR-Cas9 system: A new-fangled dawn in gene editing. Life Sci 232:116636
Gupta A, Hua L, Zhang Z, Yang B, Li W (2022) CRISPR‐induced miRNA156‐recognition element mutations in TaSPL13 improve multiple agronomic traits in wheat. Plant Biotechnol J
Hahn F, Sanjurjo Loures L, Sparks CA, Kanyuka K, Nekrasov V (2021) Efficient CRISPR/Cas-mediated targeted mutagenesis in spring and winter wheat varieties. Plants 10:1481
Hajiahmadi Z, Movahedi A, Wei H, Li D, Orooji Y, Ruan H, Zhuge Q (2019) Strategies to increase on-target and reduce off-target effects of the CRISPR/Cas9 system in plants. Int J Mol Sci 20:3719
Hao X, Song S, Zhong Q, Hajano JUD, Guo J, Wu Y (2021) Rescue of an Infectious cDNA Clone of Barley Yellow Dwarf Virus-GAV. Phytopathology 111:2383–2391
Hasan Nudin NF, van Kronenburg B, Tinnenbroek I, Krens F (2015) The importance of salicylic acid and an improved plant condition in determining success in Agrobacterium-mediated transformation. In XXV International EUCARPIA Symposium Section Ornamentals: Crossing Borders 1087:65–69
Hashemi A (2020) CRISPR–Cas9/CRISPRi tools for cell factory construction in E. coli. World J Microbiol Biotechnol 36:1–13
Hayta S, Smedley MA, Demir SU, Blundell R, Hinchliffe A, Atkinson N, Harwood WA (2019) An efficient and reproducible Agrobacterium-mediated transformation method for hexaploid wheat (Triticum aestivum L.). Plant Methods 15:1–15
Hayta S, Smedley MA, Clarke M, Forner M, Harwood WA (2021) An efficient Agrobacterium-mediated transformation protocol for hexaploid and tetraploid wheat. Curr Protoc 1:e58
He Y, Zhu M, Wang L, Wu J, Wang Q, Wang R, Zhao Y (2018) Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol Plant 11:1210–1213
He F, Wang C, Sun H, Tian S, Zhao G, Liu C, Wan C, Guo J, Huang X, Zhan G, Yu X, Kang Z, Guo J (2022) Simultaneous editing of three homoeologs of TaCIPK14 confers broad-spectrum resistance to stripe rust in wheat. Plant Biotechnol J 21:354–368
Hesami M, Jones AMP (2021) Modeling and optimizing callus growth and development in Cannabis sativa using random forest and support vector machine in combination with a genetic algorithm. Appl Microbiol Biotechnol 105:5201–5212
Hiei Y, Komari T, Kubo T (1997) Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol Biol 35:205–218
Hillary VE, Ceasar SA (2023) A Review on the Mechanism and Applications of CRISPR/Cas9/Cas12/Cas13/Cas14 Proteins Utilized for Genome Engineering. Mol Biotechnol 65:311–325
Hu J, Yu M, Chang Y, Tang H, Wang W, Du L, Wang K, Yan Y, Ye X (2022) Functional analysis of TaPDI genes on storage protein accumulation by CRISPR/Cas9 edited wheat mutants. Int J Biol Macromol 196:131–143
Ibrahim S, Saleem B, Rehman N, Zafar SA, Naeem MK, Khan MR (2022) CRISPR/Cas9 mediated disruption of Inositol Pentakisphosphate 2-Kinase 1 (TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains. J Adv Res 37:33–41
Ilhan E, Kasapoğlu AG, Muslu S, Macit M, Sezer B, Mevlütoğulları A, Guler D, Aydın M, Eksi F, Aydın M (2021) CRISPR/Cas9 and its Application in Plant Biotechnology. Nat pro Biotech 1:118–143
Isaacson W (2021) The Code Breaker: Jennifer Doudna, Gene Editing, and the Future of the Human Race, Simon & Schuster (New York, USA)
Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T (1996) High efficiency transformation of maize (Zea mays) mediated by Agrobacterium tumefaciens. Nat Biotechnol 4:745–750
Ishida Y, Tsunashima M, Hiei Y, Komari T (2015) Wheat (Triticum aestivum L.) transformation using immature embryos. Agrobacterium Protocols: 1:189–198
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakatura A (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isoenzyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169:5429–5433
Jangra A, Chaturvedi S, Kumar N, Singh H, Sharma V, Thakur M, Tiwari S, Chhokar V (2022) Polyamines: The Gleam of Next-Generation Plant Growth Regulators for Growth, Development, Stress Mitigation, and Hormonal Crosstalk in Plants—A Systematic Review. J Plant Growth Regul 1–25
Janik E, Niemcewicz M, Ceremuga M, Krzowski L, Saluk-Bijak J, Bijak M (2020) Various aspects of a gene editing system—crispr–cas9. Int J Mol Sci 21:9604
Jarvis P (2020) Environmental technology for the sustainable development goals (SDGs). Environ Technol 41:2155–2156
Jayavaradhan R, Pillis DM, Goodman M, Zhang F, Zhang Y, Andreassen PR, Malik P (2019) CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat Commun 10:2866
Jiang F, Doudna JA (2017) CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys 46:505–529
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013a) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41:e188
Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013b) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Jones HD, Doherty A, Wu H (2005) Review of methodologies and a protocol for the Agrobacterium-mediated transformation of wheat. Plant Methods 1:1–9
Jouanin A, Borm T, Boyd LA, Cockram J, Leigh F, Santos BA, Visser RGF, Smulders MJM (2019) Development of the GlutEnSeq capture system for sequencing gluten gene families in hexaploid bread wheat with deletions or mutations induced by γ-irradiation or CRISPR/Cas9. J Cereal Sci 88:157–166
Kapusi E, Stoger E (2022) Molecular farming in seed crops: gene transfer into barley (Hordeum vulgare) and wheat (Triticum aestivum). Methods Mol Biol 2480:49–60
Karakas FP, Keskin CN, Agil F, Zencirci N (2022) Phenolic composition and antioxidant potential in Turkish einkorn, emmer, durum, and bread wheat grain and grass. S Afr J Bot 149:407–415
Ke W, Shi L, Liang X, Zhao P, Wang W, Liu J, Chang Y, Hiei Y, Yanagihara C, Du P, Ishida Y, Guo Y (2021) The gene TaCB1 overcomes genotype dependency in wheat genetic transformation
Khanna H, Daggard G (2003) Agrobacterium tumefaciens-mediated transformation of wheat using a superbinary vector and a polyamine-supplemented regeneration medium. Plant Cell Rep 21:429–436
Kharb P, Chaudhary R, Tuteja N, Kaushik P (2022) A Genotype-Independent, Simple, Effective and Efficient in Planta Agrobacterium-Mediated Genetic Transformation Protocol. Methods Protoc 5:69
Khurana J, Chugh A, Khurana P (2002) Regeneration from mature and immature embryos and transient gene expression via Agrobacterium mediated transformation in emmer wheat (Triticum diccocum Schuble). Indian J Exp Biol 40:1295–1303
Kim KH, Kim JY (2021) Understanding Wheat Starch Metabolism in Properties, Environmental Stress Condition, and Molecular Approaches for Value-Added Utilization. Plants 10:2282
Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S, Kim JH, Kim JH, Kim JS (2017) In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun 8:1–12
Kim D, Alptekin B, Budak H (2018) CRISPR/Cas9 genome editing in wheat. Funct Integr Genom 18:31–41
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK (2016) High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529:490–495
Koonin EV, Makarova KS, Zhang F (2017) Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37:67–78
Kou H, Zhang Z, Yang Y, Wei C, Xu L, Zhang G (2023) Advances in the Mining of Disease Resistance Genes from Aegilops tauschii and the Utilization in Wheat. Plants 12:880
Kuluev BR, Mikhailova EV, Kuluev AR, Galimova AA, Zaikina EA, Khlestkina EK (2022) Genome Editing in Species of the Tribe Triticeae with the CRISPR/Cas System. Mol Biol 56:885–901
Kumar V, Jain M (2015) The CRISPR-Cas system for plant genome editing: Advances and opportunities. J Exp Bot 66:47–57
Kumar R, Mamrutha HM, Kaur A, Grewal A (2017a) Synergistic effect of cefotaxime and timentin to suppress the Agrobacterium overgrowth in wheat (Triticum aestivum L.) transformation. Asian J Microbiol Biotechnol Environm Sci 19:961–967
Kumar A, Lal MK, Kar SS, Nayak L, Ngangkham U, Samantaray S, Sharma SG (2017b) Bioavailability of iron and zinc as affected by phytic acid content in rice grain. J Food Biochem 41:e12413
Kumar R, Mamrutha HM, Kaur A, Venkatesh K, Sharma D, Singh GP (2019) Optimization of Agrobacterium-mediated transformation in spring bread wheat using mature and immature embryos. Mol Biol Rep 46:1845–1853
Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E (2016) CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res 44:W272–W276
Langner T, Kamoun S, Belhaj K (2018) CRISPR crops: plant genome editing toward disease resistance. Annu Rev Phytopathol 56:479–512
Lawrenson T, Shorinola O, Stacey N, Li C, Østergaard L, Patron N, Uauy C, Harwood W (2015) Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol 16:258
Lee K, Zhang Y, Kleinstiver BP, Guo JA, Aryee MJ, Miller J, Malzahn A, Zarecor S, Lawrence-Dill CJ, Joung JK, Qi Y, Wang K (2019) Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol J 17:362–372
Lee BR, La VH, Park SH, Mamun MA, Bae DW, Kim TH (2022) Dimethylthiourea Alleviates Drought Stress by Suppressing Hydrogen Peroxide-Dependent Abscisic Acid-Mediated Oxidative Responses in an Antagonistic Interaction with Salicylic Acid in Brassica napus Leaves. Antioxidants 11:2283
Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7:1494–1496
Li X, Krasnyanski SF, Korban SS (2002) Optimization of the uidA gene transfer into somatic embryos of rose via Agrobacterium tumefaciens. Plant Physiol Biochem 40:453–459
Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31:688–691
Li GL, Quan R, Wang HQ, Ruan XF, Mo JX, Zhong CL, Yang HQ, Li ZC, Gu T, Liu D, Wu ZF, Cai GY, Zhang XW (2019a) Inhibition of KU70 and KU80 by CRISPR interference, not NgAgo interference, increases the efficiency of homologous recombination in pig fetal fibroblasts. J Integr Agric 18:438–448
Li J, Li Y, Ma L (2019b) CRISPR/Cas9-based genome editing and its applications for functional genomic analyses in plants. Small Methods 3:1800473
Li J, Wang Z, He G, Ma L, Deng XW (2020) CRISPR/Cas9-mediated disruption of TaNP1 genes results in complete male sterility in bread wheat. J Genet Genom 47:263–272
Li J, Jiao G, Sun Y, Chen J, Zhong Y, Yan L, Jiang D, Ma Y, Xia L (2021a) Modification of starch composition, structure and properties through editing of TaSBEIIa in both winter and spring wheat varieties by CRISPR/Cas9. Plant Biotechnol J 19:937–951
Li J, Li Y, Ma L (2021) Recent advances in CRISPR/Cas9 and applications for wheat functional genomics and breeding. aBIOTECH 2:1–11
Li S, Lin D, Zhang Y, Deng M, Chen Y, Lv B, Li B, Lei Y, Wang Y, Zhao L, Liang Y, Liu J, Chen K, Liu Z, Xiao J, Qiu JL, Gao C (2022) Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602:455–460
Liang X, Bie X, Qiu Y, Wang K, Yang Z, Jia Y, Xu Z, Yu M, Du L, Lin Z, Ye X (2022) Development of powdery mildew resistant derivatives of wheat variety Fielder for use in genetic transformation. Crop J
Liu L, Fan XD (2014) CRISPR–Cas system: a powerful tool for genome engineering. Plant Mol Biol 85:209–218
Liu H, Wei Z, Dominguez A, Li Y, Wang X, Qi LS (2015) CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics 31:3676–3678
Liu X, Wu S, Xu J, Sui C, Wei J (2017) Application of CRISPR/Cas9 in plant biology. Acta Pharm Sin B 7:292–302
Liu J, Nannas NJ, Fu FF, Shi J, Aspinwall B, Parrott WA, Dawe RK (2019) Genome-scale sequence disruption following biolistic transformation in rice and maize. Plant Cell 31:368–383
Liu C, Zhong Y, Qi X, Chen M, Liu Z, Chen C, Tian X, Li J, Jiao Y, Wang D, Wang Y, Li M, Xin M, Liu W, Jin W, Chen S (2020a) Extension of the in vivo haploid induction system from diploid maize to hexaploid wheat. Plant Biotechnol J 18:316
Liu H, Wang K, Jia Z, Gong Q, Lin Z, Du L, Pei X, Ye X (2020b) Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J Exp Bot 71:1337–1349
Liu Q, Yang F, Zhang J, Liu H, Rahman S, Islam S, Ma W, She M (2021) Application of CRISPR/Cas9 in crop quality improvement. Int J Mol Sci 22:4206
Liu H, Chen W, Li Y, Sun L, Chai Y, Chen H, Huang C (2022) CRISPR/Cas9 technology and its utility for crop improvement. Int J Mol Sci 23:10442
Liu D, Yang H, Zhang Z, Chen Q, Guo W, Rossi V, Xin M, Du J, Hu Z, Liu J, Peng H, Ni Z, Sun Q, Yao Y (2023) An elite γ-gliadin allele improves end-use quality in wheat. New Phytol. https://doi.org/10.1111/nph.18722
Loureiro A, da Silva GJ (2019) Crispr-cas: Converting a bacterial defence mechanism into a state-of-the-art genetic manipulation tool. Antibiotics 8:18
Lv J, Yu K, Wei J, Gui H, Liu C, Liang D, Wang Y, Zhou H, Carlin R, Rich R, Lu T, Que Q, Wang WC, Zhang X, Kelliher T (2020) Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3. Nat Biotechnol 38:1397–1401
Ma C, Ha K, Kim MS, Noh YW, Lin H, Tang L, Chen H, Han S, Zhang P (2018) The anaphase promoting complex promotes NHEJ repair through stabilizing Ku80 at DNA damage sites. Cell Cycle 17:1138–1145
Maharajan T, Krishna TP, Rakkammal K, Ceasar SA, Ramesh M (2022) Application of CRISPR/Cas system in cereal improvement for biotic and abiotic stress tolerance. Planta 256:1–17
Mahfouz MM, Piatek A, Stewart CN Jr (2014) Genome engineering via TALENs and CRISPR/Cas9 systems: challenges and perspectives. Plant Biotechnol J 12:1006–1014
Mahler M, Costa AR, van Beljouw SP, Fineran PC, Brouns SJ (2022) Approaches for bacteriophage genome engineering. Trends Biotechnol. https://doi.org/10.1016/j.tibtech.2022.08.008
Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV (2006) A putative RNA-interferencebased immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1:7
Makarova KS, Wolf YI, Alkhnbashi OS et al (2015) An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol 13:722–736
Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, Moineau S, Mojica FJM, Scott D, Shah SA, Siksnys V, Terns MP, Venclovas Č, White MF, Yakunin AF, Yan W, Zhang F, Garrett RA, Backofen R, van der Oost J, Barrangou R, Koonin EV (2020) Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18:67–83
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Sci 339:823–826
Manghwar H, Li B, Ding X, Hussain A, Lindsey K, Zhang X, Jin S (2020) CRISPR/Cas systems in genome editing: methodologies and tools for sgRNA design, off-target evaluation, and strategies to mitigate off-target effects. Adv Sci 7:1902312
Marraffini LA, Sontheimer EJ (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845
Marwein R, Debbarma J, Sarki YN, Baruah I, Saikia B, Boruah HPD, Velmurugan N, Chikkaputtaiah C, (2019) Genetic engineering/genome editing approaches to modulate signaling processes in abiotic stress tolerance. Plant Signal Mol s 63–82. Woodhead Publishing
McLaughlin JE, Darwish NI, Garcia-Sanchez J, Tyagi N, Trick HN, McCormick S, Macky RD, Tumer NE (2021) A lipid transfer protein has antifungal and antioxidant activity and suppresses Fusarium head blight disease and DON accumulation in transgenic wheat. Phytopathology 111:671–683
Mühleisen J, Piepho HP, Maurer HP, Longin CFH, Reif JC (2014) Yield stability of hybrids versus lines in wheat, barley, and triticale. Theor Appl Genet 127:309–316
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Mushke R, Yarra R, Kirti PB (2019) Improved salinity tolerance and growth performance in transgenic sunflower plants via ectopic expression of a wheat antiporter gene (TaNHX2). Mol Biol Rep 46:5941–5953
Nasir A, Caetano-Anollés G (2015) A phylogenomic data-driven exploration of viral origins and evolution. Sci Adv 1:e1500527
Nassar M, Nassar R, Maki H, Al-Yagoob A, Hachim M, Senok A, Williams D, Hiraishi N (2021) Phytic acid: Properties and potential applications in dentistry. Front Mater 8:638909
Natalini A, Acciarri N, Cardi T (2021) Breeding for nutritional and organoleptic quality in vegetable crops: The case of tomato and cauliflower. Agriculture 11:606
Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31:691–693
Nerkar G, Devarumath S, Purankar M, Kumar A, Valarmathi R, Devarumath R, Appunu C (2022) Advances in crop breeding through precision genome editing. Front Genet 13:880195
Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata S, Dohmae N, Ishitani R, Zhang F, Nureki O (2015) Crystal Structure of Cas9 in Complex with gRNA and target DNA. 156:935–949.
Nonaka S, Yuhashi KI, Takada K, Sugaware M, Minamisawa K, Ezura H (2008) Ethylene production in plants during transformation suppresses vir gene expression in Agrobacterium tumefaciens. New Phytol 178:647–656
Nussenzweig PM, Marraffini LA (2020) Molecular mechanisms of CRISPR-Cas immunity in bacteria. Annu Rev Genet 93–120
Ober ES, Alahmad S, Cockram J, Forestan C, Hickey LT, Kant J, Watt M (2021) Wheat root systems as a breeding target for climate resilience. Theor Appl Genet 134:1645–1662
O’Connell MR (2019) Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR–Cas systems. J Mol Biol 431:66–87
Okada A, Arndell T, Borisjuk N, Sharma N, Watson-Haigh NS, Tucker EJ, Baumann U, Langridge P, Whitford R (2019) CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol J 17:1905–1913
Opabode JT (2006) Agrobacterium-mediated transformation of plants: emerging factors that influence efficiency. Biotechnol Mol Biol Rev 1:12–20
Ozturk A, Erdem E, Aydin M, Karaoglu MM (2022) The effects of drought after anthesis on the grain quality of bread wheat depend on drought severity and drought resistance of the variety. Cereal Res Commun 50:105–116
Park DH, Mirabella R, Bronstein PA, Preston GM, Haring MA, Lim CK, Collmer A, Schuurink RC (2010) Mutations in γ-aminobutyric acid (GABA) transaminase genes in plants or Pseudomonas syringae reduce bacterial virulence. Plant J 64:318–330
Park J, Bae S, Kim JS (2015) Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31:4014–4016
Parmar SS, Jaiwal PK, Agarwal N, Kaushik SK (2015) Optimization and validation of Agrobacterium-mediated genetic transformation for commercial indian bread wheat (Triticum aestivum L.) Cultivars using mature embryo. Cell Tissue Res 15:5301
Parry MA, Madgwick PJ, Bayon C, Tearall K, Hernandez-Lopez A, Baudo M, Rakszegi M, Hamada W, Al-Yassin A, Ouabbou H, Labhilili M, Phillips AL (2009) Mutation discovery for crop improvement. J Exp Bot 60:2817–2825
Partier A, Gay G, Tassy C, Beckert M, Feuillet C, Barret P (2017) Molecular and FISH analyses of a 53-kbp intact DNA fragment inserted by biolistics in wheat (Triticum aestivum L.) genome. Plant Cell Rep 36:1547–1559
Pellegrineschi A, Noguera LM, Skovmand B, Brito RM, Velazquez L, Salgado MM, Hernandez R, Warburton M, Hoisington D (2002) Identification of highly transformable wheat genotypes for mass production of fertile transgenic plants. Genome 45:421–430
Pereira R, Oliveira J, Sousa M (2020) Bioinformatics and computational tools for next-generation sequencing analysis in clinical genetics. J Clin Med 9:132
Perez Rojo F, Nyman RKM, Johnson AAT, Navarro P, Ryan MH, Erskine W, Kaur P (2018) CRISPR-Cas systems: ushering in the new genome editing era. Bioengineered 9:214–221
Pérez-Piñeiro P, Gago J, Landín M, Gallego PP (2012) Agrobacterium-mediated transformation of wheat: general overview and new approaches to model and identify the key factors involved. Transgenic Plants-Advances and Limitations. Rijeka, Croatia: Intech Open Access Publisher 326
Pickar-Oliver A, Gersbach CA (2019) The next generation of CRISPR–Cas technologies and applications. Nat Rev Mol 20:490–507
Pinilla-Redondo R, Mayo-Muñoz D, Russel J, Garrett RA, Randau L, Sørensen SJ, Shah SA (2020) Type IV CRISPR–Cas systems are highly diverse and involved in competition between plasmids. Nucleic Acids Res 48:2000–2012
Prykhozhij SV, Rajan V, Gaston D, Berman JN (2015) Correction: CRISPR MultiTargeter: a web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences. PLoS ONE 10:e0138634
Puren H, Reddy BJ, Sarma A, Singh SK, Ansari WA (2023) Molecular Approaches for Biofortification of Cereal Crops. In Biofortification in Cereals: Progress and Prospects 21–58 Singapore: Springer Nature Singapore
Qin G, Wu S, Zhang L, Li Y, Liu C, Yu J, Deng L, Xiao G, Zhang Z (2022) An Efficient Modular Gateway Recombinase-Based Gene Stacking System for Generating Multi-Trait Transgenic Plants. Plants 11:488
Qiu F, Xing S, Xue C, Liu J, Chen K, Chai T, Gao C (2022) Transient expression of a TaGRF4-TaGIF1 complex stimulates wheat regeneration and improves genome editing. Sci China Life Sci 65:731–738
Que Q, Elumalai S, Li X, Zhong H, Nalapalli S, Schweiner M, Fei X, Nuccio M, Kelliher T, Gu W, Chen Z, Chilton MDM (2014) Maize transformation technology development for commercial event generation. Front Plant Sci 5:279
Raffan S, Sparks C, Huttly A, Hyde L, Martignago D, Mead A, Hanley SJ, Wilkinson PA, Barker G, Edwards KJ, Curtis TY, Usher S, Kosik O, Halford NG (2021) Wheat with greatly reduced accumulation of free asparagine in the grain, produced by CRISPR/Cas9 editing of asparagine synthetase gene TaASN2. Plant Biotechnol J 19:1602–1613
Raman V, Rojas CM, Vasudevan B, Dunning K, Kolape J, Oh S, Yun J, Yang L, Li G, Pant BD, Jiang Q, Mysore KS (2022) Agrobacterium expressing a type III secretion system delivers Pseudomonas effectors into plant cells to enhance transformation. Nat Commun 13:1–14
Ramesh P, Mallikarjuna G, Sameena S, Kumar A, Gurulakshmi K, Reddy BV, Reddy PCO, Sekhar AC (2020) Advancements in molecular marker technologies and their applications in diversity studies. J Biosci. https://doi.org/10.1007/s12038-020-00089-4
Rao MJ, Wang L (2021) CRISPR/Cas9 technology for improving agronomic traits and future prospective in agriculture. Planta 254:1–16
Reegan AD, Ceasar SA, Paulraj MG, Ignacimuthu S, Al-Dhabi NA (2016) Current status of genome editing in vector mosquitoes: a review. Biosci Trends 10:424–432
Ren Q, Sretenovic S, Liu S, Tang X, Huang L, He Y, Liu L, Guo Y, Zhong Z, Liu G, Cheng Y, Zheng X, Pan C, Yin D, Zhang Y, Li W, Qi L, Li C, Qi Y, Zhang Y (2021) PAM-less plant genome editing using a CRISPR–SpRY toolbox. Nat Plants 7:25–33
Riesenberg S, Maricic T (2018) Targeting repair pathways with small molecules increase precise genome editing in pluripotent stem cells. Nat Commun 9:2164
Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell 171:470–480
Rozov SM, Permyakova NV, Deineko EV (2019) The problem of the low rates of CRISPR/Cas9-mediated knock-ins in plants: approaches and solutions. Int J Mol Sci 20:3371
Russel J, Pinilla-Redondo R, Mayo-Muñoz D, Shah SA, Sørensen SJ (2020) CRISPRCasTyper: automated identification, annotation, and classification of CRISPR-Cas Loci. CRISPR J 3:462–469
Ryu SM, Hur JW, Kim K (2019) Evolution of CRISPR towards accurate and efficient mammal genome engineering. BMB Rep 52:475
Sabooni N, Gharaghani A (2022) Induced polyploidy deeply influences reproductive life cycles, related phytochemical features, and phytohormonal activities in blackberry species. Front Plant Sci. https://doi.org/10.3389/fpls.2022.938284
Saeed S, Usman B, Shim SH, Khan SU, Nizamuddin S, Saeed S, Shoaib Y, Jeon JS, Jung KH (2022) CRISPR/Cas-mediated editing of cis-regulatory elements for crop improvement. Plant Sci 324:111435
Sahu PK, Sao R, Mondal S, Vishwakarma G, Gupta SK, Kumar V, Sing S, Sharma D, Das BK (2020) Next generation sequencing based forward genetic approaches for identification and mapping of causal mutations in crop plants: A comprehensive review. Plants 9:1355
Salas M, Park S, Srivatanakul M, Smith R (2001) Temperature influence on stable T-DNA integration in plant cells. Plant Cell Rep 20:701–705
Samson JE, Magadan AH, Moineau S (2015) The CRISPR-Cas immune system and genetic transfers: reaching an equilibrium. Microbiol Spectr 3:3–1
Sánchez-León S, Gil-Humanes J, Ozuna CV, Giménez MJ, Sousa C, Voytas DF, Barro F (2018) Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol J 16:902–910
Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V (2011) The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res 39:9275–9282
Sarkhel S, Roy A (2022) Phytic acid and its reduction in pulse matrix: Structure–function relationship owing to bioavailability enhancement of micronutrients. J Food Process Eng 45:e14030
Savadi S, Prasad P, Kashyap PL, Bhardwaj SC (2018) Molecular breeding technologies and strategies for rust resistance in wheat (Triticum aestivum) for sustained food security. Plant Pathol 67:771–791
Scheben A, Wolter F, Batley J, Puchta H, Edwards D (2017) Towards CRISPR/Cas crops–bringing together genomics and genome editing. New Phytolog 216:682–698
Schindele P, Wolter F, Puchta H (2018) Transforming plant biology and breeding with CRISPR/Cas9, Cas12 and Cas13. FEBS Lett 592:1954–1967
Schnable PS, Springer NM (2013) Progress toward understanding heterosis in crop plants. Annu Rev Plant Biol 64:71–88
Scholz I, Lange SJ, Hein S, Hess WR, Backofen R (2013) CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PloS One 8:e56470
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Jeff J, Qiu JL, Gao C (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31:686–688
Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Koonin EV (2017) Diversity and evolution of class 2 CRISPR–Cas systems. Nat Rev Microbiol 15:169–182
Shrawat AK, Lörz H (2006) Agrobacterium-mediated transformation of cereals: a promising approach crossing barriers. Plant Biotechnol J 4:575–603
Shrawat AK, Becker D, Lörz H (2007) Agrobacterium tumefaciens-mediated genetic transformation of barley (Hordeum vulgare L.). Plant Sci 172:281–290
Shreni Agrawal ER (2022) A Review: Agrobacterium-mediated gene transformation to increase plant productivity. J Phytopharmacol https://doi.org/10.31254/phyto.2022.11211
Shrivastav M, De Haro LP, Nickoloff JA (2008) Regulation of DNA double-strand break repair pathway choice. Cell Res 18:134–147
Singh P, Kumar K (2022) Agrobacterium-mediated In-planta transformation of bread wheat (Triticum aestivum L.). J Plant Biochem Biotechnol 31:206–212
Singh M, Kumar M, Albertsen MC, Young JK, Cigan AM (2018) Concurrent modifications in the three homeologs of Ms45 gene with CRISPR-Cas9 lead to rapid generation of male sterile bread wheat (Triticum aestivum L.). Plant Mol Biol 97:371–383
Singh P, Kumar K (2021) Agrobacterium-mediated In-planta transformation of bread wheat (Triticum aestivum L.). J Plant Biochem Biotechnol 1–7
Singha DL, Das D, Paswan RR, Chikkaputtaiah C, Kumar S (2022) Novel Approaches and Advanced Molecular Techniques for Crop Improvement. In Plant Microbe Interact 1–27. CRC Press.
Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V (2011) Cas3 is a single- stranded DNA nuclease and ATP- dependent helicase in the CRISPR/Cas immune system. The EMBO J 30:1335–1342
Soda N, Verma L, Giri J (2018) CRISPR-Cas9 based plant genome editing: Significance, opportunities and recent advances. Plant Physiol Biochem 131:2–11
Sparks CA, Jones HD (2004) Transformation of wheat by biolistics. Transgenic crops of the world: essential protocols, 19–34
Stemmer M, Thumberger T, del Sol KM, Wittbrodt J, Mateo JL (2015) CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE 10:e0124633
Stevanovic M, Piotter E, McClements ME, MacLaren RE (2022) Crispr Systems Suitable for Single Aav Vector Delivery. Curr Gene Ther 22:1–14
Sun Y, Li J, Xia L (2016) Precise genome modification via sequence-specific nucleases-mediated gene targeting for crop improvement. Front Plant Sci 7:1928
Sun W, Liu H, Yin W, Qiao J, Zhao X, Liu Y (2022) Strategies for enhancing the homology-directed repair efficiency of CRISPR-cas systems. CRISPR J 5:7–18
Tamulaitis G, Venclovas Č, Siksnys V (2017) Type III CRISPR-Cas immunity: major differences brushed aside. Trends Microbiol 25:49–61
Tănăsescu EC, Lite MC (2022) Harmful health effects of pesticides used on museum textile artifacts-overview. Ecotoxicol Environ Saf 247:114240
Tao LL, Yin GX, Du LP, Shi ZY, She MY, Xu HJ, Ye XG (2011) Improvement of plant regeneration from immature embryos of wheat infected by Agrobacterium tumefaciens. Agr Sci China 10:317–326
Taspinar MS, Aydin M, Sigmaz B, Yildirim N, Agar G (2017) Protective role of humic acids against picloram-induced genomic instability and DNA methylation in Phaseolus vulgaris. Environ Sci Pollut Res 24:22948–22953
Taspinar MS, Sigmaz B, Aydin M, Arslan E, Güleray A (2018) Alleviative role of Β-Estradiol against 2, 4-dichlorophenoxyacetic acid genotoxicity on common bean genome. Yuzuncu Yil Univ J Agric Sci 28:1–9
Thiyagarajan K, Noguera LM, Pacheco M, Govindan V, Vikram P (2002) Agrobacterium mediated transformation and deciphering SNPs in TaLr67 gene homeologs for gene editing in wheat. bioRxiv 2022–03
Tian X, Qin Z, Zhao Y, Wen J, Lan T, Zhang L, Wang F, Qin D, Yu K, Zhao A, Hu Z, Yao Y, Ni Z, Sun Q, Smet ID, Peng H, Xin M (2022) Stress granule-associated TaMBF1c confers thermotolerance through regulating specific mRNA translation in wheat (Triticum aestivum). New Phytol 233:1719–1731
Tran TN, Sanan-Mishra N (2015) Effect of antibiotics on callus regeneration during transformation of IR 64 rice. Biotechnol Rep 7:143–149
Turhan S, Taspinar MS, Yigider E, Aydin M, Agar G (2021) The role of long terminal repeat (LTR) responses to drought in selenium-treated wheat. Environ Eng Manag J 20
Twyman RM, Christou P (2004) Plant transformation technology: particle bombardment. Handbook of Plant Biotechnology. John Wiley & Sons Ltd, Chichester
Ullah A, Nadeem F, Nawaz A, Siddique KH, Farooq M (2022) Heat stress effects on the reproductive physiology and yield of wheat. J Agron Crop Sci 208:1–17
Uniyal AP, Mansotra K, Yadav SK, Kumar V (2019) An overview of designing and selection of sgRNAs for precise genome editing by the CRISPR-Cas9 system in plants. 3 Biotech 9:1–19
Unniyampurath U, Pilankatta R, Krishnan M (2016) RNA interference in the age of CRISPR: Will CRISPR interfere with RNAi? Int J Mol Sci 17:291
Upadhyay SK, Kumar J, Alok A, Tuli R (2013) RNA-guided genome editing for target gene mutations in wheat. G3 Genes Genom Genet 3:2233–2238
Upadhyaya NM, Mago R, Panwar V, Hewitt T, Luo M, Chen J, Sperschneider J, Nguyen-Phuc H, Wang A, Ortiz D, Hac L, Bhatt D, Li F, Zhang J, Ayliffe M, Figueroa M, Kanyuka K, Ellis JG, Dodds PN (2021) Genomics accelerated isolation of a new stem rust avirulence gene–wheat resistance gene pair. Nat Plants 7:1220–1228
Usman B, Nawaz G, Zhao N et al (2021) Programmed editing of rice (Oryza sativa l.) osspl16 gene using crispr/cas9 improves grain yield by modulating the expression of pyruvate enzymes and cell cycle proteins. Int J Mol Sci 22:1–19
United States Department of Agriculture (2022) Wheat Data. https://www.ers.usda.gov/data-products/wheat-data/ (10.03.2023)
Vasil V, Castillo AM, Fromm ME, Vasil IK (1992) Herbicide resistant fertile transgenic wheat plants obtained by microprojectile bombardment of regenerable embryogenic callus. Nat Biotechnol 10:667–674
Vats S, Bansal R, Rana N, Kumawat S, Bhatt V, Jadhav P, Kale V, Sathe A, Sonah H, Jugdaohsingh R, Sharma TR, Deshmukh R (2022) Unexplored nutritive potential of tomato to combat global malnutrition. Crit Rev Food Sci Nutr 62:1003–1034
Waheed S, Zeng L (2020) The critical role of miRNAs in regulation of flowering time and flower development. Genes 11:319
Wang YL, Xu MX, Yin GX, Tao LL, Wang DW, Ye XG (2009) Transgenic wheat plants derived from Agrobacterium-mediated transformation of mature embryo tissues. Cereal Res Commun 37:1–12
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotech 32:947–951
Wang H, La Russa M, Qi LS (2016) CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem 85:227–264
Wang S, Jin W, Wang K (2019a) Centromere histone H3-and phospholipase-mediated haploid induction in plants. Plant Methods 15:1–10
Wang W, Pan Q, Tian B, He F, Chen Y, Bai G, Trick HN, Akhunov E (2019b) Gene editing of the wheat homologs of TONNEAU 1-recruiting motif encoding gene affects grain shape and weight in wheat. Plant J 100:251–264
Wang X, Aguirre L, Rodríguez-Leal D, Hendelman A, Benoit M, Lippman ZB (2021) Dissecting cis-regulatory control of quantitative trait variation in a plant stem cell circuit. Nat Plants 7:419–427
Wang K, Shi L, Liang X, Zhao P, Wang W, Liu J, Chang Y, Hiei Y, Yanagihara C, Ishida Y, Ye X (2022a) The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat Plants 8:110–117
Wang SW, Gao C, Zheng YM, Yi L, Lu JC, Huang XY, Cai JB, Zhang PF, Cui YH, Ke AW (2022b) Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol Cancer 21:1–27
Wang Y, Du F, Wang J, Wang K, Tian C, Qi X, Jiao Y (2022c) Improving bread wheat yield through modulating an unselected AP2/ERF gene. Nat Plants 8:930–939
Waqeel J, Khan ST (2022) Microbial biofertilizers and micronutrients bioavailability: approaches to deal with zinc deficiencies. Microbial Biofertilizers and Micronutrient Availability. Springer, Cham, pp 239–297
Waseem M, Ahmad M, Saqib Naveed M, Pasha A, Hussain M, Ali Zafar S, Mujahid A, Rehman RS (2022) Abscisic acid mediated abiotic stress tolerance in plants. Asian J Crop Sci 1–17
Waters CA, Strande NT, Pryor JM, Strom CN, Mieczkowski P, Burkhalter MD, Oh S, Qaqish BF, Moore DT, Hendrickson EA, Ramsden DA (2014) The fidelity of the ligation step determines how ends are resolved during nonhomologous end joining. Nat Commun 5:1–11
Wijerathna-Yapa A, Ramtekey V, Ranawaka B, Basnet BR (2022) Applications of in vitro tissue culture technologies in breeding and genetic improvement of wheat. Plants 11:2273
Wolter F, Schindele P, Puchta H (2019) Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol 19:176
World Health Organization (2020) The state of food security and nutrition in the world 2020: transforming food systems for affordable healthy diets. Food Agric Org
Wu H, Sparks C, Amoah B, Jones HD (2003) Factors influencing successful Agrobacterium-mediated genetic transformation of wheat. Plant Cell Rep 21:659–668
Wu C, Cui K, Fahad S (2022) Heat stress decreases rice grain weight: Evidence and physiological mechanisms of heat effects prior to flowering. Int J Mol Sci 23:10922
Wulff BB, Krattinger SG (2022) The long road to engineering durable disease resistance in wheat. Curr Opin Biotechno 73:270–275
Xiao A, Cheng Z, Kong L, Zhu Z, Lin S, Gao G, Zhang B (2014) CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30:1180–1182
Xiao C, Zhang H, Xie F, Pan ZY, Qiu WM, Tong Z, Wang ZQ, He XJ, Xu YH, Sun ZH (2022) Evolution, gene expression, and protein-protein interaction analyses identify candidate CBL-CIPK signalling networks implicated in stress responses to cold and bacterial infection in citrus. BMC Plant Biol 22:1–17
Xu M, Wang Q, Wang G, Zhang X, Liu H, Jiang C (2022) Combatting Fusarium head blight: advances in molecular interactions between Fusarium graminearum and wheat. Phytopathol Res 4:1–16
Xue C, Greene EC (2021) DNA repair pathway choices in CRISPR-Cas9-mediated genome editing. Trends Genet 37:639–656
Ye X, Wang K, Liu H, Tang H, Qiu Y, Gong Q (2022) Genome Editing Toward Wheat Improvement. Genome Editing Technologies for Crop Improvement. Springer, Singapore, pp 241–269
Yeh CD, Richardson CD, Corn JE (2019) Advances in genome editing through control of DNA repair pathways. Nat Cell Biol 21:1468–1478
Yigider E, Taspinar MS, Aydin M, Agar G (2021) Humic acid effects on retrotransposon polymorphisms caused by zinc and iron in the maize (Zea mays L.) genome. Cereal Res Commun 49:193–198
Yimam YT, Zhou J, Akher SA, Zheng X, Qi Y, Zhang Y (2021) Improving a Quantitative Trait in Rice by Multigene Editing with CRISPR-Cas9. Rice Genome Engineering and Gene Editing: Methods and Protocols, 205–219
Yu Y, Wang J, Zhu ML, Wei ZM (2008) Optimization of mature embryo-based high frequency callus induction and plant regeneration from elite wheat cultivars grown in China. Plant Breed 127:249–255
Yuan ZC, Edlind MP, Liu P, Saenkham P, Banta LM, Wise AA, Ronzone E, Binns AN, Kerr K, Nester EW (2007) The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc Natl Acad Sci 104:11790–11795
Yuan WX, Yun-Mei YU, Chun-Cai HU, Zhao ZG (2017) Current Issues and Progress in the Application of CRISPR/Cas9 Technique. Biotechnol Bull 33:70–77
Zafar K, Sedeek KE, Rao GS, Khan MZ, Amin I, Kamel R, Mukhtar Z, Zafar M, Mahfouz MS, MM, (2020) Genome editing technologies for rice improvement: progress, prospects, and safety concerns. Front Genome Ed 2:5
Zarei A, Razban V, Hosseini SE, Tabei SMB (2019) Creating cell and animal models of human disease by genome editing using CRISPR/Cas9. J Gene Med 21:e3082
Zhang F (2019) Development of CRISPR-Cas systems for genome editing and beyond. Q Rev Biophys 52:e6
Zhang F, Huang Z (2021) Mechanistic insights into the versatile class II CRISPR toolbox. Trends Biochem Sci 47:433–450
Zhang YM, Zhang HM, Liu ZH, Guo XL, Li HC, Li GL, Jiang CZ, Zhang MC (2015) Inhibition of isoflavone biosynthesis enhanced T-DNA delivery in soybean by improving plant–Agrobacterium tumefaciens interaction. Plant Cell Tissue Organ Cult 121:183–193
Zhang Y, Liang Z, Zong Y, Wang Y, Liu J, Chen K, Qui JL, Gao C (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun 7:1–8
Zhang Y, Bai Y, Wu G, Zou S, Chen Y, Gao C, Tang D (2017) Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J 91:714–724
Zhang Y, Li D, Zhang D, Zhao X, Cao X, Dong L, Liu J, Chen K, Zhang H, Gao C, Wang D (2018) Analysis of the functions of Ta GW 2 homoeologs in wheat grain weight and protein content traits. Plant J 94:857–866
Zhang Z, Hua L, Gupta A, Tricoli D, Edwards KJ, Yang B, Li W (2019) Development of an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol J 17:1623–1635
Zhang J, Zhang H, Li S, Li J, Yan L, Xia L (2021a) Increasing yield potential through manipulating of an ARE1 ortholog related to nitrogen use efficiency in wheat by CRISPR/Cas9. J Integr Plant Biol 63:1649–1663
Zhang S, Shen J, Li D, Cheng Y (2021b) Strategies in the delivery of Cas9 ribonucleoprotein for CRISPR/Cas9 genome editing. Theranostics 11:614
Zhang S, Zhang R, Gao J, Song G, Li J, Li W, Qui Y, Li Y, Li G (2021c) CRISPR/Cas9-mediated genome editing for wheat grain quality improvement. Plant Biotechnol J 19:1684
Zhao TJ, Zhao SY, Chen HM, Zhao QZ, Hu ZM, Hou BK, Xia GM (2006) Transgenic wheat progeny resistant to powdery mildew generated by Agrobacterium inoculum to the basal portion of wheat seedling. Plant Cell Rep 25:1199–1204
Zheng M, Lin J, Liu X, Chu W, Li J, Gao Y, An K, Song W, Xin M, Yao Y, Peng H, Ni Z, Sun Q, Hu Z (2021) Histone acetyltransferase TaHAG1 acts as a crucial regulator to strengthen salt tolerance of hexaploid wheat. Plant Physiol 186:1951–1969
Zhi H, Zhou S, Pan W, Shang Y, Zeng Z, Zhang H (2022) The Promising Nanovectors for Gene Delivery in Plant Genome Engineering. Int J Mol Sci 23:8501
Zhu Y (2022) Advances in CRISPR/Cas9. Biomed Res Int 2022
Acknowledgements
The authors would like to thank Ataturk University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Gerhard Leubner.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yigider, E., Taspinar, M.S. & Agar, G. Advances in bread wheat production through CRISPR/Cas9 technology: a comprehensive review of quality and other aspects. Planta 258, 55 (2023). https://doi.org/10.1007/s00425-023-04199-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-023-04199-9