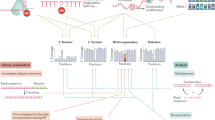
Abstract
Chemical modifications of nucleotides expand the complexity and functional properties of genomes and transcriptomes. A handful of modifications in DNA bases are part of the epigenome, wherein DNA methylation regulates chromatin structure, transcription, and co-transcriptional RNA processing. In contrast, more than 150 chemical modifications of RNA constitute the epitranscriptome. Ribonucleoside modifications comprise a diverse repertoire of chemical groups, including methylation, acetylation, deamination, isomerization, and oxidation. Such RNA modifications regulate all steps of RNA metabolism, including folding, processing, stability, transport, translation, and RNA’s intermolecular interactions. Initially thought to influence all aspects of the post-transcriptional regulation of gene expression exclusively, recent findings uncovered a crosstalk between the epitranscriptome and the epigenome. In other words, RNA modifications feedback to the epigenome to transcriptionally regulate gene expression. The epitranscriptome achieves this feat by directly or indirectly affecting chromatin structure and nuclear organization. This review highlights how chemical modifications in chromatin-associated RNAs (caRNAs) and messenger RNAs (mRNAs) encoding factors involved in transcription, chromatin structure, histone modifications, and nuclear organization affect gene expression transcriptionally.



Similar content being viewed by others
Data availability
Not applicable.
References
Abakir A, Giles TC, Cristini A et al (2020) N6-Methyladenosine regulates the stability of RNA:DNA hybrids in human cells. Nat Genet 52:48–55. https://doi.org/10.1038/s41588-019-0549-x
Abou Assi H, Rangadurai AK, Shi H et al (2021) 2′-O-Methylation can increase the abundance and lifetime of alternative RNA conformational states. Nucleic Acids Res 48:12365–12379. https://doi.org/10.1093/nar/gkaa928
Agris PF (2015) The importance of being modified: an unrealized code to RNA structure and function. RNA 21:552–554. https://doi.org/10.1261/rna.050575.115
Akichika S, Hirano S, Shichino Y et al (2019) Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363:eaav0080. https://doi.org/10.1126/SCIENCE.AAV0080
Amort T, Soulière MF, Wille A et al (2013) Long non-coding RNAs as targets for cytosine methylation. RNA Biol 10:1002–1008. https://doi.org/10.4161/rna.24454
Arango D, Sturgill D, Alhusaini N et al (2018) Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175:1872-1886.e24. https://doi.org/10.1016/j.cell.2018.10.030
Arango D, Sturgill D, Yang R et al (2022) Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine. Mol Cell 82:2797-2814.e11. https://doi.org/10.1016/J.MOLCEL.2022.05.016
Athanasiadis A, Rich A, Maas S (2004) Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2:e391. https://doi.org/10.1371/JOURNAL.PBIO.0020391
Auffinger P, Westhof E (2001) Hydrophobic groups stabilize the hydration shell of 2′-O-methylated RNA duplexes. Angewandte Chemie - Int Ed 40:4648–4650. https://doi.org/10.1002/1521-3773(20011217)40:24%3c4648::AID-ANIE4648%3e3.0.CO;2-U
Bahn JH, Lee JH, Li G et al (2012) Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res 22:142–150. https://doi.org/10.1101/GR.124107.111
Baldini L, Charpentier B, Labialle S (2021) Emerging data on the diversity of molecular mechanisms involving c/d snornas. Noncoding RNA 7:30. https://doi.org/10.3390/ncrna7020030
Banani SF, Lee HO, Hyman AA, Rosen MK (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18:285–298. https://doi.org/10.1038/nrm.2017.7
Barbieri I, Tzelepis K, Pandolfini L et al (2017) Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 552:126–131. https://doi.org/10.1038/nature24678
Belotserkovskii BP, Tornaletti S, D’Souza AD, Hanawalt PC (2018) R-loop generation during transcription: formation, processing and cellular outcomes. DNA Repair 71:69–81. https://doi.org/10.1016/j.dnarep.2018.08.009
Bizhanova A, Kaufman PD (2021) Close to the edge heterochromatin at the nucleolar and nuclear peripheries. Biochim et Biophys Acta BBA Gene Regul Mech 1864:194666. https://doi.org/10.1016/J.BBAGRM.2020.194666
Boccaletto P, Stefaniak F, Ray A et al (2022) MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res 50:D231–D235. https://doi.org/10.1093/NAR/GKAB1083
Boeren J, Gribnau J (2021) Xist-mediated chromatin changes that establish silencing of an entire X chromosome in mammals. Curr Opin Cell Biol 70:44–50. https://doi.org/10.1016/j.ceb.2020.11.004
Borchardt EK, Martinez NM, Gilbert WV (2020) Regulation and function of RNA pseudouridylation in human cells. Annu Rev Genet 54:309–336. https://doi.org/10.1146/ANNUREV-GENET-112618-043830
Boulias K, Toczydłowska-Socha D, Hawley BR et al (2019) Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Mol Cell 75:631-643.e8. https://doi.org/10.1016/j.molcel.2019.06.006
Bratkovič T, Rogelj B (2014) The many faces of small nucleolar RNAs. Biochim Biophys Acta Gene Regul Mech 1839:438–443. https://doi.org/10.1016/j.bbagrm.2014.04.009
Cattenoz PB, Taft RJ, Westhof E, Mattick JS (2013) Transcriptome-wide identification of A > I RNA editing sites by inosine specific cleavage. RNA 19:257–270. https://doi.org/10.1261/RNA.036202.112
Chaumeil J, Le Baccon P, Wutz A, Heard E (2006) A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev 20:2223–2237. https://doi.org/10.1101/gad.380906
Chelmicki T, Roger E, Teissandier A et al (2021) m6A RNA methylation regulates the fate of endogenous retroviruses. Nature 591:312–316. https://doi.org/10.1038/s41586-020-03135-1
Chen X, Li A, Sun BF et al (2019) 5-Methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nature Cell Biol 21:978–990. https://doi.org/10.1038/s41556-019-0361-y
Chen H, Yao J, Bao R et al (2021) Cross-talk of four types of RNA modification writers defines tumor microenvironment and pharmacogenomic landscape in colorectal cancer. Mol Cancer 20:1–21. https://doi.org/10.1186/S12943-021-01322-W
Cheng JX, Chen L, Li Y et al (2018) RNA cytosine methylation and methyltransferases mediate chromatin organization and 5-azacytidine response and resistance in leukaemia. Nat Commun 9:1163. https://doi.org/10.1038/s41467-018-03513-4
Chi YH, Haller K, Peloponese JM, Jeang KT (2007) Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in de-condensation of mitotic chromosomes. J Biol Chem 282:27447–27458. https://doi.org/10.1074/jbc.M703098200
Chung H, Calis JJA, Wu X et al (2018) Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown. Cell 172:811-824.e14. https://doi.org/10.1016/J.CELL.2017.12.038
Clemson CM, Hutchinson JN, Sara SA et al (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33:717–726. https://doi.org/10.1016/j.molcel.2009.01.026
Crick FHC (1966) Codon–anticodon pairing: the wobble hypothesis. J Mol Biol 19:548–555. https://doi.org/10.1016/S0022-2836(66)80022-0
Dai Q, Zhang LS, Sun HL et al (2022) Quantitative sequencing using BID-seq uncovers abundant pseudouridines in mammalian mRNA at base resolution. Nat Biotechnol 41:344-354. https://doi.org/10.1038/s41587-022-01505-w
Davidovich C, Cech TR (2015) The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA 21:2007–2022. https://doi.org/10.1093/nar/23.24.5020
Davis DR (1995) Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 23:5020–5026. https://doi.org/10.1093/nar/23.24.5020
Delorimier E, Hinman MN, Copperman J et al (2017) Pseudouridine modification inhibits muscleblind-like 1 (MBNL1) binding to CCUG repeats and minimally structured RNA through reduced RNA flexibility. J Biol Chem 292:4350–4357. https://doi.org/10.1074/jbc.M116.770768
Deng S, Zhang J, Su J et al (2022) RNA m6A regulates transcription via DNA demethylation and chromatin accessibility. Nat Genet 54:1427–1437. https://doi.org/10.1038/s41588-022-01173-1
Dimitrova DG, Teysset L, Carré C (2019) RNA 2’-O-methylation (Nm) modification in human diseases. Genes (Basel) 10:117. https://doi.org/10.3390/GENES10020117
Dominissini D, Rechavi G (2018) N4-acetylation of cytidine in mRNA by NAT10 Regulates Stability and Translation. Cell 175:1725–1727. https://doi.org/10.1016/J.CELL.2018.11.037
Dominissini D, Moshitch-Moshkovitz S, Schwartz S et al (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485:201–206. https://doi.org/10.1038/NATURE11112
Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S et al (2016) The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530:441–446. https://doi.org/10.1038/nature16998
Draycott AS, Schaening-Burgos C, Rojas-Duran MF et al (2022) Transcriptome-wide mapping reveals a diverse dihydrouridine landscape including mRNA. PLoS Biol 20:e3001622. https://doi.org/10.1371/journal.pbio.3001622
Dutta N, Deb I, Sarzynska J, Lahiri A (2022) Inosine and its methyl derivatives: occurrence, biogenesis, and function in RNA. Prog Biophys Mol Biol 169–170:21–52. https://doi.org/10.1016/J.PBIOMOLBIO.2022.01.001
Engreitz JM, Sirokman K, McDonel P et al (2014) RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell 159:188–199. https://doi.org/10.1016/j.cell.2014.08.018
Enroth C, Poulsen LD, Iversen S et al (2019) Detection of internal N7-methylguanosine (m7G) RNA modifications by mutational profiling sequencing. Nucleic Acids Res 47:E126–E126. https://doi.org/10.1093/nar/gkz736
Feric M, Vaidya N, Harmon TS et al (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165:1686–1697. https://doi.org/10.1016/J.CELL.2016.04.047
Finet O, Yague-Sanz C, Krüger LK et al (2022a) Transcription-wide mapping of dihydrouridine reveals that mRNA dihydrouridylation is required for meiotic chromosome segregation. Mol Cell 82:404-419.e9. https://doi.org/10.1016/J.MOLCEL.2021.11.003
Finet O, Yague-Sanz C, Marchand F, Hermand D (2022b) The dihydrouridine landscape from tRNA to mRNA: a perspective on synthesis, structural impact and function. RNA Biol 19:735–750. https://doi.org/10.1080/15476286.2022.2078094
Fu L, Guerrero CR, Zhong N et al (2014) Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc 136:11582–11585. https://doi.org/10.1021/JA505305Z
Ganot P, Caizergues-Ferrer M, Kiss T (1997) The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev 11(7):941–956. https://doi.org/10.1101/gad.11.7.941
Guillén-Boixet J, Kopach A, Holehouse AS et al (2020) RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181:346. https://doi.org/10.1016/J.CELL.2020.03.049
Guo G, Pan K, Fang S et al (2021) Advances in mRNA 5-methylcytosine modifications: detection, effectors, biological functions, and clinical relevance. Mol Ther Nucleic Acids 26:575–593. https://doi.org/10.1016/J.OMTN.2021.08.020
Gutschner T, Hämmerle M, Eißmann M et al (2013) The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 73:1180–1189. https://doi.org/10.1158/0008-5472.CAN-12-2850
Haag S, Sloan KE, Ranjan N et al (2016) NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J 35:2104–2119. https://doi.org/10.15252/EMBJ.201694885
Han J, Kim D, Morris KV (2007) Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA 104:12422–12427. https://doi.org/10.1073/PNAS.0701635104
Han C, Sun LY, Luo XQ et al (2022) Chromatin-associated orphan snoRNA regulates DNA damage-mediated differentiation via a non-canonical complex. Cell Rep 38:110421. https://doi.org/10.1016/j.celrep.2022.110421
He PC, Wei J, Dou X et al (2023) Exon architecture controls mRNA m6A suppression and gene expression. Science 379:677–682. https://doi.org/10.1126/science.abj9090
Helm M, Giegé R, Florentz C (1999) A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNA(Lys). Biochemistry 38:13338–13346. https://doi.org/10.1021/bi991061g
Hirose T, Ninomiya K, Nakagawa S, Yamazaki T (2022) A guide to membraneless organelles and their various roles in gene regulation. Nat Rev Mol Cell Biol 24:288–304. https://doi.org/10.1038/S41580-022-00558-8
Hsiao YHE, Bahn JH, Yang Y et al (2018) RNA editing in nascent RNA affects pre-mRNA splicing. Genome Res 28:812–823. https://doi.org/10.1101/GR.231209.117
Hsu PJ, Zhu Y, Ma H et al (2017) Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res 27:1115–1127. https://doi.org/10.1038/cr.2017.99
Huang H, Weng H, Zhou K et al (2019) Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 567:414–419. https://doi.org/10.1038/S41586-019-1016-7
Huertas P, Aguilera A (2003) Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12:711–721. https://doi.org/10.1016/j.molcel.2003.08.010
Hutchinson JN, Ensminger AW, Clemson CM et al (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8:39. https://doi.org/10.1186/1471-2164-8-39
Ito S, Akamatsu Y, Noma A et al (2014) A single acetylation of 18S rRNA is essential for biogenesis of the small ribosomal subunit in saccharomyces cerevisiae. J Biol Chem 289:26201–26212. https://doi.org/10.1074/jbc.M114.593996
Jachowicz JW, Bing X, Pontabry J et al (2017) LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat Genet 49:1502–1510. https://doi.org/10.1038/NG.3945
Jack K, Bellodi C, Landry DM et al (2011) rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell 44:660–666. https://doi.org/10.1016/J.MOLCEL.2011.09.017
Jia G, Fu Y, Zhao X et al (2011) N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7:885–887. https://doi.org/10.1038/nchembio.687
Jin H, Huo C, Zhou T, Xie S (2022) m1A RNA modification in gene expression regulation. Genes (Basel) 13:910. https://doi.org/10.3390/GENES13050910
Jobert L, Pinzón N, Van Herreweghe E et al (2009) Human U1 snRNA forms a new chromatin-associated snRNP with TAF15. EMBO Rep 10:494–500. https://doi.org/10.1038/embor.2009.24
Kan RL, Chen J, Sallam T (2022) Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet 38:182–193. https://doi.org/10.1016/j.tig.2021.06.014
Kaneko S, Bonasio R, Saldaña-Meyer R et al (2014) Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell 53:290–300. https://doi.org/10.1016/J.MOLCEL.2013.11.012
Kang HJ, Cheon NY, Park H et al (2021) TonEBP recognizes R-loops and initiates m6A RNA methylation for R-loop resolution. Nucleic Acids Res 49:269–284. https://doi.org/10.1093/nar/gkaa1162
Karthiya R, Khandelia P (2020) m6A RNA methylation: ramifications for gene expression and human health. Mol Biotechnol 62:467–484. https://doi.org/10.1007/S12033-020-00269-5
Karthiya R, Wasil SM, Khandelia P (2020) Emerging role of N4-acetylcytidine modification of RNA in gene regulation and cellular functions. Mol Biol Rep 47:9189–9199. https://doi.org/10.1007/S11033-020-05963-W
Kawai G, Yamamoto Y, Kamimura T et al (1992) Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2’-Hydroxyl Group. Biochemistry 31:1040–1046. https://doi.org/10.1021/bi00119a012
Khalil AM, Guttman M, Huarte M et al (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 106:11667–11672. https://doi.org/10.1073/PNAS.0904715106
Kierzek E, Malgowska M, Lisowiec J et al (2014) The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res 42:3492–3501. https://doi.org/10.1093/nar/gkt1330
Kim DDY, Kim TTY, Walsh T et al (2004) Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res 14:1719–1725. https://doi.org/10.1101/GR.2855504
Kim TK, Hemberg M, Gray JM et al (2010) Widespread transcription at neuronal activity-regulated enhancers. Nature 465:182–187. https://doi.org/10.1038/nature09033
Kiss-László Z, Henry Y, Bachellerie JP et al (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85:1077–1088. https://doi.org/10.1016/S0092-8674(00)81308-2
Knisbacher BA, Levanon EY (2015) DNA and RNA editing of retrotransposons accelerate mammalian genome evolution. Ann N Y Acad Sci 1341:115–125. https://doi.org/10.1111/NYAS.12713
Kovaříková AS, Bártová E (2022) PARP-dependent acetylation of N4-cytidine in RNA appears in UV-damaged chromatin. PREPRINT. Research Square. https://doi.org/10.21203/rs.3.rs-2210212/v1
Kuang W, Jin H, Yang F et al (2022) ALKBH3-dependent m1A demethylation of Aurora A mRNA inhibits ciliogenesis. Cell Discov 8:25. https://doi.org/10.1038/s41421-022-00385-3
Kumbhar BV, Kamble AD, Sonawane KD (2013) Conformational preferences of modified nucleoside N(4)-acetylcytidine, ac4C occur at “Wobble” 34th Position in the Anticodon Loop of tRNA. Cell Biochem Biophys 66:797–816. https://doi.org/10.1007/s12013-013-9525-8
Lalonde MS, Zuo Y, Zhang J et al (2007) Exoribonuclease R in Mycoplasma genitalium can carry out both RNA processing and degradative functions and is sensitive to RNA ribose methylation. RNA 13:1957–1968. https://doi.org/10.1261/rna.706207
Larrieu D, Britton S, Demir M et al (2014) Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science 344:527–532. https://doi.org/10.1126/SCIENCE.1252651
Lee JH, Wang R, Xiong F et al (2021) Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol Cell 81:3368-3385.e9. https://doi.org/10.1016/j.molcel.2021.07.024
Levanon EY, Eisenberg E, Yelin R et al (2004) Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22:1001–1005. https://doi.org/10.1038/NBT996
Lev-Maor G, Sorek R, Levanon EY et al (2007) RNA-editing-mediated exon evolution. Genome Biol 8:R29. https://doi.org/10.1186/GB-2007-8-2-R29
Li X, Fu XD (2019) Chromatin-associated RNAs as facilitators of functional genomic interactions. Nat Rev Genet 20:503–519. https://doi.org/10.1038/s41576-019-0135-1
Li Y, Syed J, Sugiyama H (2016) RNA-DNA triplex formation by long noncoding RNAs. Cell Chem Biol 23:1325–1333. https://doi.org/10.1016/j.chembiol.2016.09.011
Li X, Xiong X, Zhang M et al (2017) Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell 68:993–1005. https://doi.org/10.1016/J.MOLCEL.2017.10.019
Li Y, Xia L, Tan K et al (2020) N 6-Methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat Genet 52:870–877. https://doi.org/10.1038/s41588-020-0677-3
Li M, Tao Z, Zhao Y et al (2022) 5-Methylcytosine RNA methyltransferases and their potential roles in cancer. J Transl Med 20:214. https://doi.org/10.1186/s12967-022-03427-2
Liao L, He Y, Li SJ et al (2023) Lysine 2-hydroxyisobutyrylation of NAT10 promotes cancer metastasis in an ac4C-dependent manner. Cell Res 2023:1–17. https://doi.org/10.1038/s41422-023-00793-4
Licht K, Hartl M, Amman F et al (2019) Inosine induces context-dependent recoding and translational stalling. Nucleic Acids Res 47:3–14. https://doi.org/10.1093/NAR/GKY1163
Liu J, Dou X, Chen C et al (2020) N6-Methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367:580–586. https://doi.org/10.1126/science.aay6018
Liu J, Yue Y, Han D et al (2014) A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10:93–95. https://doi.org/10.1038/nchembio.1432
Liu N, Dai Q, Zheng G et al (2015) N6 -Methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518:560–564. https://doi.org/10.1038/nature14234
Liu F, Clark W, Luo G et al (2016) ALKBH1-mediated tRNA demethylation regulates translation. Cell 167:816-828.e16. https://doi.org/10.1016/j.cell.2016.09.038
Liu J, Gao M, He J et al (2021) The RNA m6A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature 591:322–326. https://doi.org/10.1038/s41586-021-03313-9
Liu C, Cao J, Zhang H, Yin J (2022) Evolutionary history of RNA modifications at N6-adenosine originating from the R-M system in eukaryotes and prokaryotes. Biology (basel) 11:214. https://doi.org/10.3390/BIOLOGY11020214
Lubas M, Christensen MS, Kristiansen MS et al (2011) Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 43:624–637. https://doi.org/10.1016/J.MOLCEL.2011.06.028
Lyman SK, Gerace L, Baserga SJ (1999) Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleoproteins. RNA 5:1597–1604. https://doi.org/10.1017/s1355838299991288
Blow M, Futreal PA, Wooster R, Stratton MR (2004) A survey of RNA editing in human brain. Genome Res 14:2379–2387. https://doi.org/10.1101/GR.2951204
Ma X, Yang C, Alexandrov A et al (2005) Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J 24:2403–2413. https://doi.org/10.1038/sj.emboj.7600718
Ma Y, Li W, Fan C et al (2022) Comprehensive analysis of long non-coding RNAs N4-acetylcytidine in Alzheimer’s disease mice model using high-throughput sequencing. J Alzheimers Dis 90:1659–1675. https://doi.org/10.3233/jad-220564
Mannoor K, Liao J, Jiang F (2012) Small nucleolar RNAs in cancer. Biochim Biophys Acta Rev Cancer 1826:121–128. https://doi.org/10.1016/j.bbcan.2012.03.005
Martin FH, Castro MM, Aboul-Ela F, Tinoco I (1985) Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res 13:8927–8938. https://doi.org/10.1093/NAR/13.24.8927
Mauer J, Luo X, Blanjoie A et al (2017) Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541:371–375. https://doi.org/10.1038/nature21022
Meyer KD, Saletore Y, Zumbo P et al (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149:1635–1646. https://doi.org/10.1016/j.cell.2012.05.003
Miller HE, Ilieva M, Bishop AJR, Uchida S (2022) Current status of epitranscriptomic marks affecting lncRNA structures and functions. Non-Coding RNA 8(23):8–23. https://doi.org/10.3390/NCRNA8020023
Möller-Krull M, Zemann A, Roos C et al (2008) Beyond DNA: RNA editing and steps toward Alu exonization in primates. J Mol Biol 382:601–609. https://doi.org/10.1016/J.JMB.2008.07.014
Moshitch-Moshkovitz S, Dominissini D, Rechavi G (2022) The epitranscriptome toolbox. Cell 185:764–776. https://doi.org/10.1016/j.cell.2022.02.007
Nagano T, Mitchell JA, Sanz LA et al (2008) The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322:1717–1720. https://doi.org/10.1126/SCIENCE.1163802
Ngo GHP, Grimstead JW, Baird DM (2021) UPF1 promotes the formation of R loops to stimulate DNA double-strand break repair. Nat Commun 12. https://doi.org/10.1038/s41467-021-24201-w
Ni J, Tien AL, Fournier MJ (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89:565–573. https://doi.org/10.1016/s0092-8674(00)80238-x
O’Leary VB, Ovsepian SV, Carrascosa LG et al (2015) PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep 11:474–485. https://doi.org/10.1016/j.celrep.2015.03.043
Ouvrard J, Muniz L, Nicolas E, Trouche D (2022) Small interfering RNAs targeting a chromatin-associated RNA induce its transcriptional silencing in human cells. Mol Cell Biol 42:e0027122. https://doi.org/10.1128/mcb.00271-22
Passmore LA, Coller J (2022) Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat Rev Mol Cell Biol 23:93–106. https://doi.org/10.1038/s41580-021-00417-y
Pastor WA, Aravind L, Rao A (2013) TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 14:341–356. https://doi.org/10.1038/nrm3589
Patil DP, Chen CK, Pickering BF et al (2016) M6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537:369–373. https://doi.org/10.1038/nature19342
Peng T, Hou Y, Meng H et al (2023) Mapping nucleolus-associated chromatin interactions using nucleolus Hi-C reveals pattern of heterochromatin interactions. Nature Commun 14:1–15. https://doi.org/10.1038/s41467-023-36021-1
Percharde M, Lin CJ, Yin Y et al (2018) A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 174:391-405.e19. https://doi.org/10.1016/J.CELL.2018.05.043
Ping XL, Sun BF, Wang L et al (2014) Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 24:177–189. https://doi.org/10.1038/cr.2014.3
Rashid R, Liang B, Baker DL et al (2006) Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol Cell 21:249–260. https://doi.org/10.1016/j.molcel.2005.11.017
Ries RJ, Zaccara S, Klein P et al (2019) m6A enhances the phase separation potential of mRNA. Nature 571(7765):424–428. https://doi.org/10.1038/s41586-019-1374-1
Rodriguez J, Menet JS, Rosbash M (2012) Nascent-Seq indicates widespread cotranscriptional RNA editing in Drosophila. Mol Cell 47:27–37. https://doi.org/10.1016/J.MOLCEL.2012.05.002
Rošić S, Köhler F, Erhardt S (2014) Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J Cell Biol 207:335–349. https://doi.org/10.1083/jcb.201404097
Sabari BR, Dall’Agnese A, Young RA (2020) Biomolecular condensates in the nucleus. Trends Biochem Sci 45:961–977. https://doi.org/10.1016/J.TIBS.2020.06.007
Safra M, Sas-Chen A, Nir R et al (2017) The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551:251–255. https://doi.org/10.1038/nature24456
Sahakyan A, Yang Y, Plath K (2018) The role of Xist in X-chromosome dosage compensation. Trends Cell Biol 28:999–1013. https://doi.org/10.1016/j.tcb.2018.05.005
Salas-Armenteros I, Pérez-Calero C, Bayona-Feliu A et al (2017) Human THO –Sin3A interaction reveals new mechanisms to prevent R-loops that cause genome instability. EMBO J 36:3532–3547. https://doi.org/10.15252/embj.201797208
Sasaki YTF, Ideue T, Sano M et al (2009) MEN/noncoding RNAs are essential for structural integrity of nuclear paraspeckles. PNAS 106:2525–2530. https://doi.org/10.1073/pnas.0807899106
Sas-Chen A, Thomas JM, Matzov D et al (2020) Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature 583:638–643. https://doi.org/10.1038/s41586-020-2418-2
Schmitz KM, Mayer C, Postepska A, Grummt I (2010) Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24:2264–2269. https://doi.org/10.1101/gad.590910
Selmi T, Hussain S, DIetmann S et al (2021) Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res 49:1006–1022. https://doi.org/10.1093/nar/gkaa1193
Seo KW, Kleiner RE (2020) YTHDF2 recognition of N1-methyladenosine (m1A)-modified RNA is associated with transcript destabilization. ACS Chem Biol 15:132–139. https://doi.org/10.1021/acschembio.9b00655
Sharma S, Langhendries JL, Watzinger P et al (2015) Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res 43:2242–2258. https://doi.org/10.1093/nar/gkv075
Sharma S, Poetz F, Bruer M et al (2016) Acetylation-dependent control of global poly(A) RNA degradation by CBP/p300 and HDAC1/2. Mol Cell 63:927–938. https://doi.org/10.1016/j.molcel.2016.08.030
Shen Q, Zhang Q, Shi Y et al (2018) Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature 554:123–127. https://doi.org/10.1038/nature25434
Shi H, Wei J, He C (2019) Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell 74:640–650. https://doi.org/10.1016/J.MOLCEL.2019.04.025
Shiromoto Y, Sakurai M, Minakuchi M et al (2021) ADAR1 RNA editing enzyme regulates R-loop formation and genome stability at telomeres in cancer cells. Nature Commun 12:1–18. https://doi.org/10.1038/s41467-021-21921-x
Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D (2008) Nucleolus: the fascinating nuclear body. Histochem Cell Biol 129:13–31. https://doi.org/10.1007/S00418-007-0359-6
Somme J, Van Laer B, Roovers M et al (2014) Characterization of two homologous 2′-O-methyltransferases showing different specificities for their tRNA substrates. RNA 20:1257–1271. https://doi.org/10.1261/rna.044503.114
Statello L, Guo CJ, Chen LL, Huarte M (2021) Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22:96–118. https://doi.org/10.1038/s41580-020-00315-9
Stenström L, Mahdessian D, Gnann C et al (2020) Mapping the nucleolar proteome reveals a spatiotemporal organization related to intrinsic protein disorder. Mol Syst Biol 16:e9469. https://doi.org/10.15252/MSB.20209469
Stern L, Schulman LH (1978) The role of the minor base N4-acetylcytidine in the function of the Escherichia coli noninitiator methionine transfer RNA. J Biol Chem 253:6132–6139. https://doi.org/10.1016/S0021-9258(17)34590-8
Sun Q, Hao Q, Prasanth KV (2018) Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet 34:142–157. https://doi.org/10.1016/j.tig.2017.11.005
Taniguchi T, Miyauchi K, Sakaguchi Y et al (2018) Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis. Nat Chem Biol 14:1010–1020. https://doi.org/10.1038/s41589-018-0119-z
Tan-Wong SM, Dhir S, Proudfoot NJ (2019) R-loops promote antisense transcription across the mammalian genome. Mol Cell 76:600-616.e6. https://doi.org/10.1016/J.MOLCEL.2019.10.002
Taoka M, Nobe Y, Yamaki Y et al (2018) Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res 46:9289–9298. https://doi.org/10.1093/NAR/GKY811
Tartakoff A, DiMario P, Hurt E et al (2022) The dual nature of the nucleolus. Genes Dev 36:765–769. https://doi.org/10.1101/GAD.349748.122
Thalalla Gamage S, Bortolin-Cavaillé M-L, Link C et al (2022) Antisense pairing and SNORD13 structure guide RNA cytidine acetylation. RNA 28:1582–1596. https://doi.org/10.1261/RNA.079254.122
Tomikawa C (2018) 7-Methylguanosine modifications in transfer RNA (tRNA). Int J Mol Sci 19:4080. https://doi.org/10.3390/ijms19124080
Tsai MC, Manor O, Wan Y et al (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329:689–693. https://doi.org/10.1126/SCIENCE.1192002
Tschochner H, Hurt E (2003) Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol 13:255–263. https://doi.org/10.1016/S0962-8924(03)00054-0
Uzonyi A, Dierks D, Nir R et al (2023) Exclusion of m6A from splice-site proximal regions by the exon junction complex dictates m6A topologies and mRNA stability. Mol Cell 83:237–251.e7. https://doi.org/10.1016/j.molcel.2022.12.026
Vaidyanathan PP, Alsadhan I, Merriman DK et al (2017) Pseudouridine and N6-methyladenosine modifications weaken PUF protein/RNA interactions. RNA 23:611–618. https://doi.org/10.1261/rna.060053.116
Voigts-Hoffmann F, Hengesbach M, Kobitski AY et al (2007) A methyl group controls conformational equilibrium in human mitochondrial tRNA Lys. J AM CHEM SOC 129:13382–13383. https://doi.org/10.1021/ja075520
Wada T, Takagi T, Yamaguchi Y et al (1998) DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 12:343–356. https://doi.org/10.1101/GAD.12.3.343
Wang S, Kool ET (1995) Origins of the large differences in stability of DNA and RNA helices: C-5 methyl and 2’-hydroxyl effects. Biochemistry 34:4125–4132. https://doi.org/10.1021/BI00012A031
Wang X, Lu Z, Gomez A et al (2014) N6-Methyladenosine-dependent regulation of messenger RNA stability. Nature 505:117–120. https://doi.org/10.1038/nature12730
Wang Y, Li Y, Yue M et al (2018) N6-Methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci 21:195–206. https://doi.org/10.1038/s41593-017-0057-1
Wang Z, Wang X, Zhang T et al (2021) LncRNA MALAT1 promotes gastric cancer progression via inhibiting autophagic flux and inducing fibroblast activation. Cell Death Dis 12:1–17. https://doi.org/10.1038/s41419-021-03645-4
Warda AS, Kretschmer J, Hackert P et al (2017) Human METTL16 is a N 6 -methyladenosine (m 6 A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep 18:2004–2014. https://doi.org/10.15252/embr.201744940
Wei J, Liu F, Lu Z et al (2018) Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell 71:973–985.e5. https://doi.org/10.1016/j.molcel.2018.08.011
Wu Y, Chen Z, Xie G et al (2022) RNA m1A methylation regulates glycolysis of cancer cells through modulating ATP5D. Proc Natl Acad Sci USA 119:e2119038119. https://doi.org/10.1073/PNAS.2119038119
Wu C, Chen W, He J et al (2020) Interplay of m6A and H3K27 trimethylation restrains inflammation during bacterial infection. Sci Adv 6:eaba0647. https://doi.org/10.1126/sciadv.aba0647
Wurm AA, Pina C (2019) Long non-coding RNAs as functional and structural chromatin modulators in acute myeloid leukemia. Front Oncol 9:899. https://doi.org/10.3389/fonc.2019.00899
Xia X, Wang Y, Zheng JC (2023) Internal m7G methylation: a novel epitranscriptomic contributor in brain development and diseases. Mol Ther Nucleic Acids 31:295–308. https://doi.org/10.1016/J.OMTN.2023.01.003
Xiang Y, Laurent B, Hsu CH et al (2017) RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 543:573–576. https://doi.org/10.1038/nature21671
Xiao W, Adhikari S, Dahal U et al (2016) Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol Cell 61:507–519. https://doi.org/10.1016/j.molcel.2016.01.012
Xiao YL, Liu S, Ge R et al (2023) Transcriptome-wide profiling and quantification of N 6-methyladenosine by enzyme-assisted adenosine deamination. Nat Biotechnol. https://doi.org/10.1038/s41587-022-01587-6
Xie Y, Castro-Hernández R, Sokpor G et al (2019) RBM15 modulates the function of chromatin remodeling factor BAF155 through RNA methylation in developing cortex. Mol Neurobiol 56:7305–7320. https://doi.org/10.1007/s12035-019-1595-1
Xu C, Wang X, Liu K et al (2014) Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 10:927–929. https://doi.org/10.1038/nchembio.1654
Xu W, Li J, He C et al (2021) METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature 591:317–321. https://doi.org/10.1038/s41586-021-03210-1
Xu W, He C, Kaye EG et al (2022) Dynamic control of chromatin-associated m6A methylation regulates nascent RNA synthesis. Mol Cell 82:1156-1168.e7. https://doi.org/10.1016/j.molcel.2022.02.006
Yang L, Lin C, Liu W et al (2011) NcRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147:773–788. https://doi.org/10.1016/j.cell.2011.08.054
Yang X, Yang Y, Sun BF et al (2017) 5-Methylcytosine promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res 27:606–625. https://doi.org/10.1038/cr.2017.55
Yang F, Tanasa B, Micheletti R et al (2021) Shape of promoter antisense RNAs regulates ligand-induced transcription activation. Nature 595:444–449. https://doi.org/10.1038/s41586-021-03589-x
Yu F, Tanaka Y, Yamashita K et al (2011) Molecular basis of dihydrouridine formation on tRNA. PNAS 108:19593–19598. https://doi.org/10.1073/pnas.1112352108
Zhang LS, Liu C, Ma H et al (2019) Transcriptome-wide mapping of internal N7-methylguanosine methylome in mammalian mRNA. Mol Cell 74:1304-1316.e8. https://doi.org/10.1016/j.molcel.2019.03.036
Zhang C, Chen L, Peng D et al (2020) METTL3 and N6-methyladenosine promote homologous recombination-mediated repair of DSBs by modulating DNA-RNA hybrid accumulation. Mol Cell 79:425-442.e7. https://doi.org/10.1016/j.molcel.2020.06.017
Zhang LS, Xiong QP, Peña Perez S et al (2021) ALKBH7-mediated demethylation regulates mitochondrial polycistronic RNA processing. Nat Cell Biol 23:684–691. https://doi.org/10.1038/S41556-021-00709-7
Zhang L, Bernardo KD, Vickers TA et al (2022) NAT10 and DDX21 Proteins interact with RNase H1 and affect the performance of phosphorothioate oligonucleotides. Nucleic Acid Ther 32:280–299. https://doi.org/10.1089/nat.2021.0107
Zhao J, Sun BK, Erwin JA et al (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322:750–756. https://doi.org/10.1126/SCIENCE.1163045
Zhao X, Patton JR, Davis SL et al (2004) Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol Cell 5:549–58. https://doi.org/10.1016/j.molcel.2004.06.044
Zheng G, Dahl JA, Niu Y et al (2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49:18–29. https://doi.org/10.1016/j.molcel.2012.10.015
Zheng Q, Gan H, Yang F et al (2020) Cytoplasmic m1A reader YTHDF3 inhibits trophoblast invasion by downregulation of m1A-methylated IGF1R. Cell Discov 6:12. https://doi.org/10.1038/s41421-020-0144-4
Zhou KI, Parisien M, Dai Q et al (2016) N6-Methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J Mol Biol 428:822–833. https://doi.org/10.1016/j.jmb.2015.08.021
Acknowledgements
We thank Kevin Vasquez for insightful discussions.
Funding
This work was supported by the National Cancer Institute (NCI) grant R00CA245035, the American Association for Cancer Research (AACR) grant 22–20-01-ARAN, and a V Scholar Award V2022-016. E.A.P. was funded by the Austrian Marshall Plan Foundation.
Author information
Authors and Affiliations
Contributions
E.A.P., S.R., and D.A. wrote the first draft. E.A.P. and D.A. edited the final draft. D.A. prepared the figures.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Patrasso, E.A., Raikundalia, S. & Arango, D. Regulation of the epigenome through RNA modifications. Chromosoma 132, 231–246 (2023). https://doi.org/10.1007/s00412-023-00794-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-023-00794-7