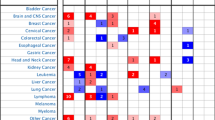
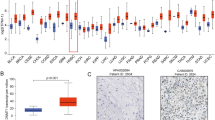
Abstract
Objective
Dysregulation of RNA binding proteins (RBPs) plays an important role in controlling processes in cancer development. However, the function of RBPs has not been thoroughly and systematically documented in head and neck cancer. We aim to explore the role of RPB in the pathogenesis of HNSC.
Methods
We obtained HNSC gene expression data and corresponding clinical information from The Cancer Genome Atlas (TCGA) and the GEO databases, and identified aberrantly expressed RBPs between tumors and normal tissues. Meanwhile, we performed a series of bioinformatics to explore the function and prognostic value of these RBPs.
Results
A total of 249 abnormally expressed RBPs were identified, including 101 downregulated RBPs and 148 upregulated RBPs. Using least absolute shrinkage and selection operator (LASSO) and univariate Cox regression analysis, the 15 RPBs were identified as hub genes. With the 15 RPBS, the prognostic prediction model was constructed. Further analysis showed that the high-risk score of the patients expressed a better survival outcome. The prediction model was validated in another HNSC dataset, and similar findings were observed.
Conclusions
Our results provide novel insights into the pathogenesis of HNSC. The fifteen RBP gene signature exhibited the predictive value of moderate HNSC prognosis, and have potential application value in clinical decision-making and individualized treatment.










Similar content being viewed by others
Availability of data and material
Data availability could be obtained from TCGA website.
References
Preciado DA, Matas A, Adams GL (2002) Squamous cell carcinoma of the head and neck in solid organ transplant recipients. Head Neck 24:319–325
Patel BP, Shah PM, Rawal UM, Desai AA, Shah SV, Rawal RM, Patel PS (2005) Activation of MMP-2 and MMP-9 in patients with oral squamous cell carcinoma. J Surg Oncol 90:81–88
Choi KK, Kim MJ, Yun PY, Lee JH, Moon HS, Lee TR, Myoung H (2006) Independent prognostic factors of 861 cases of oral squamous cell carcinoma in Korean adults. Oral Oncol 42:208–217
Wozniak A, Szyfter K, Szyfter W, Florek E (2012) Head and neck cancer–history. Przegl Lek 69:1079–1083
Mehanna H, Paleri V, West CM, Nutting C (2010) Head and neck cancer–part 1: epidemiology, presentation, and prevention. BMJ 341:c4684
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68:7–30
Chi AC, Day TA, Neville BW (2015) Oral cavity and oropharyngeal squamous cell carcinoma—an update. CA Cancer J Clin 65:401–421
Gerstberger S, Hafner M, Tuschl T (2014) A census of human RNA-binding proteins. Nat Rev Genet 15:829–845
Velasco MX, Kosti A, Penalva LOF, Hernandez G (2019) The diverse roles of RNA-binding proteins in glioma development. Adv Exp Med Biol 1157:29–39
Fredericks AM, Cygan KJ, Brown BA, Fairbrother WG (2015) RNA-binding proteins: splicing factors and disease. Biomolecules 5:893–909
Suzuki A, Niimi Y, Shinmyozu K, Zhou Z, Kiso M, Saga Y (2016) Dead end1 is an essential partner of NANOS2 for selective binding of target RNAs in male germ cell development. EMBO Rep 17:37–46
Chang ET, Parekh PR, Yang Q, Nguyen DM, Carrier F (2016) Heterogenous ribonucleoprotein A18 (hnRNP A18) promotes tumor growth by increasing protein translation of selected transcripts in cancer cells. Oncotarget 7:10578–10593
Chenard CA, Richard S (2008) New implications for the QUAKING RNA binding protein in human disease. J Neurosci Res 86:233–242
Neelamraju Y, Gonzalez-Perez A, Bhat-Nakshatri P, Nakshatri H, Janga SC (2018) Mutational landscape of RNA-binding proteins in human cancers. RNA Biol 15:115–129
Wichmann G, Rosolowski M, Krohn K, Kreuz M, Boehm A, Reiche A, Scharrer U, Halama D, Bertolini J, Bauer U et al (2015) The role of HPV RNA transcription, immune response-related gene expression and disruptive TP53 mutations in diagnostic and prognostic profiling of head and neck cancer. Int J Cancer 137:2846–2857
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287
Wu TT, Chen YF, Hastie T, Sobel E, Lange K (2009) Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics 25:714–721
Thul PJ, Lindskog C (2018) The human protein atlas: a spatial map of the human proteome. Protein Sci 27:233–244
Lan Y, Xiao X, He Z, Luo Y, Wu C, Li L, Song X (2018) Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res 46:5809–5821
Dong W, Dai ZH, Liu FC, Guo XG, Ge CM, Ding J, Liu H, Yang F (2019) The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine 45:155–167
Soni S, Anand P, Padwad YS (2019) MAPKAPK2: the master regulator of RNA-binding proteins modulates transcript stability and tumor progression. J Exp Clin Cancer Res 38:121
Schultz CW, Preet R, Dhir T, Dixon DA, Brody JR (2020) Understanding and targeting the disease-related RNA binding protein human antigen R (HuR). Wiley Interdiscip Rev RNA 11:e1581
Frisone P, Pradella D, Di Matteo A, Belloni E, Ghigna C, Paronetto MP (2015) SAM68: signal transduction and RNA metabolism in human cancer. Biomed Res Int 2015:528954
Perron G, Jandaghi P, Solanki S, Safisamghabadi M, Storoz C, Karimzadeh M, Papadakis AI, Arseneault M, Scelo G, Banks RE et al (2018) A general framework for interrogation of mRNA stability programs identifies RNA-binding proteins that govern cancer transcriptomes. Cell Rep 23:1639–1650
Wurth L (2012) Versatility of RNA-binding proteins in cancer. Comp Funct Genomics 2012:178525
Lukong KE, Chang KW, Khandjian EW, Richard S (2008) RNA-binding proteins in human genetic disease. Trends Genet 24:416–425
Zhang H, Brown RD, Stenmark KR, Hu CJ (2020) RNA-binding proteins in pulmonary hypertension. Int J Mol Sci 21:3757
Zhu S, Rooney S, Michlewski G (2020) RNA-targeted therapies and high-throughput screening methods. Int J Mol Sci 21:2996
Wong KK (2020) DNMT1 as a therapeutic target in pancreatic cancer: mechanisms and clinical implications. Cell Oncol (Dordr) 43:779–792
Kah KW (2020) DNMT1: a key drug target in triple-negative breast cancer. Semin Cancer Biol 72:198–213
Wu L, Gong Y, Yan T, Zhang H (2020) LINP1 promotes the progression of cervical cancer by scaffolding EZH2, LSD1 and DNMT1 to inhibit the expression of KLF2 and PRSS8. Biochem Cell Biol 98:591–599
Guo T, Zambo KDA, Zamuner FT, Ou T, Hopkins C, Kelley DZ, Wulf HA, Winkler E, Erbe R, Danilova L et al (2020) Chromatin structure regulates cancer-specific alternative splicing events in primary HPV-related oropharyngeal squamous cell carcinoma. Epigenetics 15:1–13
Li J, Feng D, Gao C, Zhang Y, Xu J, Wu M, Zhan X (2019) Isoforms S and L of MRPL33 from alternative splicing have isoformspecific roles in the chemoresponse to epirubicin in gastric cancer cells via the PI3K/AKT signaling pathway. Int J Oncol 54:1591–1600
Liu L, Luo C, Luo Y, Chen L, Liu Y, Wang Y, Han J, Zhang Y, Wei N, Xie Z et al (2018) MRPL33 and its splicing regulator hnRNPK are required for mitochondria function and implicated in tumor progression. Oncogene 37:86–94
Hua WF, Zhong Q, Xia TL, Chen Q, Zhang MY, Zhou AJ, Tu ZW, Qu C, Li MZ, Xia YF et al (2016) RBM24 suppresses cancer progression by upregulating miR-25 to target MALAT1 in nasopharyngeal carcinoma. Cell Death Dis 7:e2352
Yang SW, Li L, Connelly JP, Porter SN, Kodali K, Gan H, Park JM, Tacer KF, Tillman H, Peng J et al (2020) A cancer-specific ubiquitin ligase drives mRNA alternative polyadenylation by ubiquitinating the mRNA 3’ end processing complex. Mol Cell 77(1206–1221):e1207
Lu M, Ge Q, Wang G, Luo Y, Wang X, Jiang W, Liu X, Wu CL, Xiao Y, Wang X (2018) CIRBP is a novel oncogene in human bladder cancer inducing expression of HIF-1alpha. Cell Death Dis 9:1046
Sun D, Wang Y, Wang H, Xin Y (2020) The novel long non-coding RNA LATS2-AS1-001 inhibits gastric cancer progression by regulating the LATS2/YAP1 signaling pathway via binding to EZH2. Cancer Cell Int 20:204
Funding
The works were supported by Guizhou Science and Technology Department (Identification of Slug-regulated miRNA in oral squamous cell carcinoma and its mechanism of tumor metastasis inhibition, NO: Qiankehe LH [2014] No. 7012).
Author information
Authors and Affiliations
Contributions
JKS, XFD, XLC wrote the main manuscript text. JKS, KZ, XLC prepared Figs. 1–9. JKS, XFD contributed to data analysis. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Duan, X., Cheng, X., Yin, X. et al. Systematic analysis of the function and prognostic value of RNA binding protein in head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol 279, 1535–1547 (2022). https://doi.org/10.1007/s00405-021-06929-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-021-06929-9