Abstract
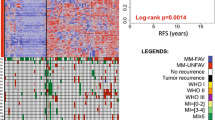
DNA methylation patterns delineate clinically relevant subgroups of meningioma. We previously established the six meningioma methylation classes (MC) benign 1–3, intermediate A and B, and malignant. Here, we set out to identify subgroup-specific mutational patterns and gene regulation. Whole genome sequencing was performed on 62 samples across all MCs and WHO grades from 62 patients with matched blood control, including 40 sporadic meningiomas and 22 meningiomas arising after radiation (Mrad). RNA sequencing was added for 18 of these cases and chromatin-immunoprecipitation for histone H3 lysine 27 acetylation (H3K27ac) followed by sequencing (ChIP-seq) for 16 samples. Besides the known mutations in meningioma, structural variants were found as the mechanism of NF2 inactivation in a small subset (5%) of sporadic meningiomas, similar to previous reports for Mrad. Aberrations of DMD were found to be enriched in MCs with NF2 mutations, and DMD was among the most differentially upregulated genes in NF2 mutant compared to NF2 wild-type cases. The mutational signature AC3, which has been associated with defects in homologous recombination repair (HRR), was detected in both sporadic meningioma and Mrad, but widely distributed across the genome in sporadic cases and enriched near genomic breakpoints in Mrad. Compared to the other MCs, the number of single nucleotide variants matching the AC3 pattern was significantly higher in the malignant MC, which also exhibited higher genomic instability, determined by the numbers of both large segments affected by copy number alterations and breakpoints between large segments. ChIP-seq analysis for H3K27ac revealed a specific activation of genes regulated by the transcription factor FOXM1 in the malignant MC. This analysis also revealed a super enhancer near the HOXD gene cluster in this MC, which, together with general upregulation of HOX genes in the malignant MC, indicates a role of HOX genes in meningioma aggressiveness. This data elucidates the biological mechanisms rendering different epigenetic subgroups of meningiomas, and suggests leveraging HRR as a novel therapeutic target.





Similar content being viewed by others
References
Abedalthagafi MS, Bi WL, Merrill PH et al (2015) ARID1A and TERT promoter mutations in dedifferentiated meningioma. Cancer Genet 208:345–350
Abkevich V, Timms KM, Hennessy BT et al (2012) Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer 107:1776–1782
Alexandrov LB, Nik-Zainal S, Wedge DC et al (2013) Signatures of mutational processes in human cancer. Nature 500:415–421
Birkbak NJ, Wang ZC, Kim JY et al (2012) Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov 2:366–375
Brastianos PK, Horowitz PM, Santagata S et al (2013) Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet 45:285–289
Carroll TS, Liang Z, Salama R, Stark R, de Santiago I (2014) Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front Genet 5:75
Chen X, Muller GA, Quaas M et al (2013) The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol Cell Biol 33:227–236
Chudasama P, Mughal SS, Sanders MA et al (2018) Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat Commun 9:144
Clark VE, Erson-Omay EZ, Serin A et al (2013) Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science 339:1077–1080
Clark VE, Harmanci AS, Bai H et al (2016) Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet 48:1253–1259
Collord G, Tarpey P, Kurbatova N et al (2018) An integrated genomic analysis of anaplastic meningioma identifies prognostic molecular signatures. Sci Rep 8:13537
Di Vinci A, Brigati C, Casciano I et al (2012) HOXA7, 9, and 10 are methylation targets associated with aggressive behavior in meningiomas. Transl Res 160:355–362
Engert F, Kovac M, Baumhoer D, Nathrath M, Fulda S (2017) Osteosarcoma cells with genetic signatures of BRCAness are susceptible to the PARP inhibitor talazoparib alone or in combination with chemotherapeutics. Oncotarget 8:48794–48806
Engert F, Schneider C, Weibeta LM, Probst M, Fulda S (2015) PARP inhibitors sensitize Ewing sarcoma cells to temozolomide-induced apoptosis via the mitochondrial pathway. Mol Cancer Ther 14:2818–2830
Gallia GL, Zhang M, Ning Y, Haffner MC, Batista D, Binder ZA (2018) Genomic analysis identifies frequent deletions of Dystrophin in olfactory neuroblastoma. Nat Commun. 9(1):5410
Gao F, Shi L, Russin J et al (2013) DNA methylation in the malignant transformation of meningiomas. PLoS One 8:e54114
Georgiou G, van Heeringen SJ (2016) fluff: exploratory analysis and visualization of high-throughput sequencing data. PeerJ 4:e2209
Godlewski J, Kiezun J, Krazinski BE, Kozielec Z, Wierzbicki PM, Kmiec Z (2018) The immunoexpression of YAP1 and LATS1 proteins in clear cell renal cell carcinoma: impact on patients’ survival. Biomed Res Int 2018:2653623
Gu Z, Eils R, Schlesner M (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849
Harmanci AS, Youngblood MW, Clark VE et al (2017) Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nat Commun 8:14433
Heining C, Horak P, Uhrig S et al (2018) NRG1 fusions in KRAS wild-type pancreatic cancer. Cancer Discov 8:1087–1095
Helmrich A, Ballarino M, Tora L (2011) Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell 44:966–977
Jiang Y, Qiu Y, Minn AJ, Zhang NR (2016) Assessing intratumor heterogeneity and tracking longitudinal and spatial clonal evolutionary history by next-generation sequencing. Proc Natl Acad Sci USA 113:E5528–5537
Jones DT, Hutter B, Jager N et al (2013) Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet 45:927–932
Juratli TA, McCabe D, Nayyar N et al (2018) DMD genomic deletions characterize a subset of progressive/higher-grade meningiomas with poor outcome. Acta Neuropathol 136:779–792
Juratli TA, Thiede C, Koerner MVA et al (2017) Intratumoral heterogeneity and TERT promoter mutations in progressive/higher-grade meningiomas. Oncotarget 8:109228–109237
Khan A, Zhang X (2016) dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res 44:D164–171
Kishida Y, Natsume A, Kondo Y et al (2012) Epigenetic subclassification of meningiomas based on genome-wide DNA methylation analyses. Carcinogenesis 33:436–441
Kovac M, Blattmann C, Ribi S et al (2015) Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat Commun 6:8940
Kulakovskiy IV, Vorontsov IE, Yevshin IS et al (2018) HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res 46:D252–D259
Laurendeau I, Ferrer M, Garrido D et al (2010) Gene expression profiling of the hedgehog signaling pathway in human meningiomas. Mol Med 16:262–270
Lawrence MS, Stojanov P, Polak P, Kryukov GV et al (2013) Mutational heterogenieity in cancer and the search for new cancer genes. Nature. https://doi.org/10.1038/nature12213
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Loven J, Hoke HA, Lin CY et al (2013) Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153:320–334
Mahaney BL, Meek K, Lees-Miller SP (2009) Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J 417:639–650
McLean CY, Bristor D, Hiller M et al (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28:495–501
Oh JE, Ohta T, Satomi K et al (2015) Alterations in the NF2/LATS1/LATS2/YAP pathway in Schwannomas. J Neuropathol Exp Neurol 74:952–959
Olar A, Wani KM, Wilson CD et al (2017) Global epigenetic profiling identifies methylation subgroups associated with recurrence-free survival in meningioma. Acta Neuropathol 133:431–444
Popova T, Manie E, Rieunier G et al (2012) Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res 72:5454–5462
Rehrauer H, Wu L, Blum W et al (2018) How asbestos drives the tissue towards tumors: YAP activation, macrophage and mesothelial precursor recruitment, RNA editing, and somatic mutations. Oncogene 37:2645–2659
Reuss DE, Piro RM, Jones DT et al (2013) Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol 125:351–358
Ross-Innes CS, Stark R, Teschendorff AE et al (2012) Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481:389–393
Rubio-Perez C, Tamborero D, Schroeder MP, Antolín AA, Deu-Pons J, Perez-Llamas C et al (2015) In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals novel targeting opportunities. Cancer Cell 27(3):382–96
Sahm F, Schrimpf D, Olar A et al (2016) TERT promoter mutations and risk of recurrence in meningioma. J Natl Cancer Inst 108:djv377
Sahm F, Schrimpf D, Stichel D et al (2017) DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol 18:682–694
Sahm F, Toprak UH, Hubschmann D et al (2017) Meningiomas induced by low-dose radiation carry structural variants of NF2 and a distinct mutational signature. Acta Neuropathol 134:155–158
Smith MJ, O’Sullivan J, Bhaskar SS et al (2013) Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet 45:295–298
Spiegl-Kreinecker S, Lotsch D, Neumayer K et al (2018) TERT promoter mutations are associated with poor prognosis and cell immortalization in meningioma. Neuro Oncol 20:1584–1593
Sturm D, Orr BA, Toprak UH et al (2016) New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164:1060–1072
Telli ML, Timms KM, Reid J et al (2016) Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res 22:3764–3773
Vasudevan HN, Braunstein SE, Phillips JJ et al (2018) Comprehensive molecular profiling identifies FOXM1 as a key transcription factor for meningioma proliferation. Cell Rep 22:3672–3683
Whyte WA, Orlando DA, Hnisz D et al (2013) Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153:307–319
Zhang Y, Liu T, Meyer CA et al (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137
Acknowledgements
This project was supported by the German Cancer Aid (110983, 70112007), the Else Kröner-Fresenius Stiftung (2015_A060, 2017_EKES.24), and the Heidelberg Center for Personalized Oncology (DKFZ-HIPO). We further thank the DKFZ Omics IT and Data Management Core Facility (ODCF) and DKFZ Genomics and Proteomics Core Facility for technical support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (XLSX 10 kb)
Online Resource 1: Supplementary Table 1 Cohort characteristics
Supplementary material 2 (XLSX 138 kb)
Online Resource 2: Supplementary Table 2 Functional exonic variants and small insertions/deletions.
Supplementary material 3 (XLSX 6 kb)
Online Resource 3: Supplementary Table 3 Association of methylation classes with functional exonic small variants
Supplementary material 4 (PDF 394 kb)
Online Resource 4: Supplementary Table 4 Quality control metrics of 16 ChIP-seq samples generated using the ChIPQC R package. Supplementary Figure 1 Volcano plot showing differential expression in NF2 mutant vs wild-type cases. Supplementary Figure 2 Mutational signature analysis (legend below figure). Supplementary Figure 3 Correlation between genomic instability and AC3 stratified for Mrad and sporadic cases. Supplementary Figure 4 HOX gene expression
Supplementary material 5 (CSV 15731 kb)
Online Resource 5 Expression data from RNA sequencing from FFPE samples
Supplementary material 6 (CSV 12660 kb)
Online Resource 6 Expression data from RNA sequencing from frozen samples
Rights and permissions
About this article
Cite this article
Paramasivam, N., Hübschmann, D., Toprak, U.H. et al. Mutational patterns and regulatory networks in epigenetic subgroups of meningioma. Acta Neuropathol 138, 295–308 (2019). https://doi.org/10.1007/s00401-019-02008-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-019-02008-w