Abstract
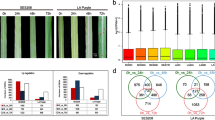
The beneficial interactions between plant and endophytes can enormously improve plant resistance against phytopathogens. Endophytic bacteria are well known for their plant propitious attributes. A tobacco seed endophyte Bacillus amyloliquefaciens YN201732 can enhance the ability of tobacco plants to resist powdery mildew, but the mechanisms operating at molecular level are highly elusive. In this study, transcriptome of YN201732-treated and non-treated tobacco leaves were compared under pathogen challenge and control conditions. Overall, analysis elucidates that DEGs related to biosynthesis of secondary metabolites, plant hormone signal transduction, phenylpropanoid biosynthesis, flavonoid biosynthesis, metabolic pathways, immune system process, and disease resistance-related biological functions were enriched in endophyte-treated diseased plant. The key genes of JA/ET pathway were significantly upregulated in tobacco plant after endophyte YN201732 treatment, which indicates the activation of JA/ET-mediated disease resistance against powdery mildew. Additionally, chitinase gene and synthesis of lignin and flavonoids were increased in diseased plant upon endophyte treatment which could help plant to resist pathogen infection. In conclusion, the study provides the molecular dissection of B. amyloliquefaciens YN201732-induced resistance in tobacco plant against powdery mildew and lays a theoretical foundation of employing endophytic bacteria against plant diseases.





Similar content being viewed by others
Data Availability
All the data are present inside manuscript file. Raw sequencing data have been deposited to the Sequence Read Archive (SRA) under BioProject PRJNA825699.
References
Abdallah RAB, Stedel C, Garagounis C, Nefzi A, Jabnoun-Khiareddine H, Papadopoulou KK, Daami-Remadi M (2017) Involvement of lipopeptide antibiotics and chitinase genes and induction of host defense in suppression of Fusarium wilt by endophytic Bacillus spp. in tomato. Crop Prot 99:45–58
Agarwal A, Kaul V, Faggian R, Rookes JE, Ludwig-Müller J, Cahill DM (2011) Analysis of global host gene expression during the primary phase of the Arabidopsis thaliana–Plasmodiophora brassicae interaction. Funct Plant Biol 38:462–478
Agarwal H, Dowarah B, Baruah PM, Bordoloi KS, Krishnatreya DB, Agarwala N (2020) Endophytes from Gnetum gnemon L. can protect seedlings against the infection of phytopathogenic bacterium Ralstonia solanacearum as well as promote plant growth in tomato. Microbiol Res. https://doi.org/10.1016/j.micres.2020.126503
Ahmed A, Munir S, He P, Li Y, He P, Yixin W, He Y (2020) Biocontrol arsenals of bacterial endophyte: an imminent triumph against clubroot disease. Microbiol Res. https://doi.org/10.1016/j.micres.2020.126565
Ahmed W, Zhou G, Yang J, Munir S, Ahmed A, Liu Q, Zhao Z, Ji G (2022) Bacillus amyloliquefaciens WS-1 ṁ0 as a potential plant growth-promoter and biocontrol agent for bacterial wilt disease of flue-cured tobacco. Egypt J Biol Pest Cont 32:1–14
Araújo WL, Marcon J, Maccheroni W Jr, Van Elsas JD, Van Vuurde JW, Azevedo JL (2002) Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol 68:4906–4914
Arce-Leal ÁP, Bautista R, Rodríguez-Negrete EA, Manzanilla-Ramírez MÁ, Velázquez-Monreal JJ, Santos-Cervantes ME, Méndez-Lozano J, Beuzón CR, Bejarano ER, Castillo AG (2020) Gene expression profile of mexican lime (Citrus aurantifolia) trees in response to huanglongbing disease caused by Candidatus liberibacter asiaticus. Microorganisms 8:528
Aswani R, Thomas R, Radhakrishnan E (2022) "Induction of plant defense response by endophytic microorganisms. Biocontrol Mechanisms of Endophytic Microorganisms. Elsevier, Amsterdam, pp 89–115
Barkodia M, Joshi U, Rami N, Wati L (2018) Endophytes: a hidden treasure inside plant. IJCS 6:1660–1665
Belkhadir Y, Subramaniam R, Dangl JL (2004) Plant disease resistance protein signaling: NBS–LRR proteins and their partners. Curr Opin Plant Biol 7:391–399
Bodhankar S, Grover M, Hemanth S, Reddy G, Rasul S, Yadav SK, Desai S, Mallappa M, Mandapaka M, Srinivasarao C (2017) Maize seed endophytic bacteria: dominance of antagonistic, lytic enzyme-producing Bacillus spp. 3 Biotech. https://doi.org/10.1007/s13205-017-0860-0
Chakraborty N, Chakraborty N, Acharyya P, Acharya K (2021) Isolation, characterization and identification of novel broad spectrum bacterial antagonist (s) to control Fusarium wilt of eggplant. Physiol Mol Plant Pathol 116:101711
Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21:2527–2540
Choudhary DK, Johri BN (2009) Interactions of Bacillus spp. and plants–with special reference to induced systemic resistance (ISR). Microbiol Res 164:493–513
Cui K, Wu W-W, Diao Q-Y (2019) Application and research progress on transcriptomics. Biotechnol Bull 35:1
Devendar P, Qu R-Y, Kang W-M, He B, Yang G-F (2018) Palladium-catalyzed cross-coupling reactions: a powerful tool for the synthesis of agrochemicals. J Agric Food Chem 66:8914–8934
Dietz J (2007) Recently introduced powdery mildew fungicides. In: Krmer W, Schirmer U (eds) Modern Crop Protection Compounds. Wiley-VCH Verlag GmbH, Germany
Dong X, Zhao Y, Ran X, Guo L, Zhao D-G (2017) Overexpression of a new chitinase gene EuCHIT2 enhances resistance to Erysiphe cichoracearum DC in tobacco plants. Int J Mol Sci. https://doi.org/10.3390/ijms18112361
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10:366–371
Fouda A, Hassan SED, Eid AM, El-Din Ewais E (2019) The interaction between plants and bacterial endophytes under salinity stress. In: Endophytes and secondary metabolites. Springer, Cham, pp 591–607
Griffin MR (2014) Biocontrol and bioremediation: two areas of endophytic research which hold great promise. Advances in Endophytic Research. Springer, Cham, pp 257–282
Gu Y-Q, Wildermuth MC, Chakravarthy S, Loh Y-T, Yang C, He X, Han Y, Martin GB (2002) Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14:817–831
Hermosa R, Rubio MB, Cardoza RE, Nicolás C, Monte E, Gutiérrez S (2013) The contribution of Trichoderma to balancing the costs of plant growth and defense. Int Microbiol 16:69–80
Hu H, Wang C, Li X, Tang Y, Wang Y, Chen S, Yan S (2018) RNA-Seq identification of candidate defense genes targeted by endophytic Bacillus cereus-mediated induced systemic resistance against Meloidogyne incognita in tomato. Pest Manag Sci 74:2793–2805
Jasim B, Sreelakshmi K, Mathew J, Radhakrishnan E (2016) Surfactin, iturin, and fengycin biosynthesis by endophytic Bacillus sp. from Bacopa monnieri. Microb Ecol 72:106–119
Jia H, Zhao P, Wang B, Tariq P, Zhao F, Zhao M, Wang Q, Yang T, Fang J (2016) Overexpression of polyphenol oxidase gene in strawberry fruit delays the fungus infection process. Plant Mol Biol Report 34:592–606
Jiao R, Liu J, Yang H, He P, Wu Y, Wang J, Wang G, He Y (2018) Isolation and identification of endophytic bacteria inhibiting Phytophthora parasitica var. nicotianae and promoting tobacco seedling growth. J Yunnan Agric Univ 33:1037–1045
Jiao R, Munir S, He P, Yang H, Wu Y, Wang J, He P, Cai Y, Wang G, He Y (2020) Biocontrol potential of the endophytic Bacillus amyloliquefaciens YN201732 against tobacco powdery mildew and its growth promotion. Biol Control 143:104160
Kaul S, Sharma T, Dhar K, M. (2016) “Omics” tools for better understanding the plant–endophyte interactions. Front Plant Sci 7:955
Kim J, Lee SB, Suh MC (2021) Arabidopsis 3-ketoacyl-CoA synthase 4 is essential for root and pollen tube growth. J Plant Biol 64:155–165
Kumar D (2014) Salicylic acid signaling in disease resistance. Plant Sci 228:127–134
La Cruz-López D, Cruz-López L, Holguín-Meléndez F, Guillén-Navarro GK, Huerta-Palacios G (2022) Volatile organic compounds produced by cacao endophytic bacteria and their inhibitory activity on Moniliophthora roreri. Curr Microbiol 79:1–11
Lee MH, Jeon HS, Kim SH, Chung JH, Roppolo D, Lee HJ, Cho HJ, Tobimatsu Y, Ralph J, Park OK (2019) Lignin-based barrier restricts pathogens to the infection site and confers resistance in plants. EMBO J 38:e101948
Lewandowska M, Keyl A, Feussner I (2020) Wax biosynthesis in response to danger: Its regulation upon abiotic and biotic stress. New Phytol 227:698–713
Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16:319–331
Li L, Long Y, Li H, Wu X (2020) Comparative transcriptome analysis reveals key pathways and hub genes in rapeseed during the early stage of Plasmodiophora brassicae infection. Front Genet 10:1275
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:1–21
Ma X, Keller B, Mcdonald BA, Palma-Guerrero J, Wicker T (2018) Comparative transcriptomics reveals how wheat responds to infection by Zymoseptoria tritici. Mol Plant Microbe Interact 31:420–431
Malik A (2019) Purification and properties of plant chitinases: a review. J Food Biochem 43:e12762
Mayo S, Cominelli E, Sparvoli F, González-López O, Rodríguez-González A, Gutiérrez S, Casquero PA (2016) Development of a qPCR strategy to select bean genes involved in plant defense response and regulated by the Trichoderma velutinum–Rhizoctonia solani interaction. Front Plant Sci 7:1109
Mishra A, Singh SP, Mahfooz S, Singh SP, Bhattacharya A, Mishra N, Nautiyal C (2018) Endophyte-mediated modulation of defense-related genes and systemic resistance in Withania somnifera (L.) Dunal under Alternaria alternata stress. Appl Environ Microbiol. https://doi.org/10.1016/j.bbagrm.2011.08.004
Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) AP2/ERF family transcription factors in plant abiotic stress responses. Biochimica Et Biophysica Acta -Gene Regul Mech 1819:86–96
Munir, S, Ahmed, N, Abid M, Rehman SU, Ashraf M, Zhang L, Anees M (2019) Chitinolytic activity of the indigenous Trichoderma spp. from the north west of Pakistan against the fungal phytopathogens. Pak J Bot 51:711–716. https://doi.org/10.30848/PJB2019-2(37)
Pichersky E, Raguso RA (2018) Why do plants produce so many terpenoid compounds? New Phytol 220:692–702
Pozo MJ, Van Der Ent S, Van Loon L, Pieterse CM (2008) Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol 180:511–523
Qi W, Xiaojie C, Shuangyue G, Xinyue Z, Tianlu H, Ting D (2019) Transcriptome profiling of maize resistance gene in response to DZSY21 induction. Acta Agric Zhejiangensis 31:345
Ruan J, Zhou Y, Zhou M, Yan J, Khurshid M, Weng W, Cheng J, Zhang K (2019) Jasmonic acid signaling pathway in plants. Int J Mol Sci 20:2479
Safara S, Harighi B, Bahramnejad B, Ahmadi S (2022) Antibacterial activity of endophytic bacteria against sugar beet root rot agent by volatile organic compound production and induction of systemic resistance. Front Microbiol. https://doi.org/10.3389/fmicb.2022.921762
Sahu PK, Singh S, Gupta A, Singh UB, Brahmaprakash G, Saxena AK (2019) Antagonistic potential of bacterial endophytes and induction of systemic resistance against collar rot pathogen Sclerotium rolfsii in tomato. Biol Control 137:104014
Sandhya V, Ali SZ (2018) Quantitative mRNA analysis of induced genes in maize inoculated with Acinetobacter baumannii Strain MZ30V92. Curr Biotechnol 7:438–452
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Shetty R, Fretté X, Jensen B, Shetty NP, Jensen JD, Jørgensen HJL, Newman M-A, Christensen LP (2011) Silicon-induced changes in antifungal phenolic acids, flavonoids, and key phenylpropanoid pathway genes during the interaction between miniature roses and the biotrophic pathogen Podosphaera pannosa. Plant Physiol 157:2194–2205
Singh SP, Gaur R (2017) Endophytic Streptomyces spp. underscore induction of defense regulatory genes and confers resistance against Sclerotium rolfsii in chickpea. Biol Cont 104:44–56
Singh KB, Foley RC, Oñate-Sánchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5:430–436
Sun D, Liao J, Sun L, Wang Y, Liu Y, Deng Q, Zhang N, Xu D, Fang Z, Wang W, Gooneratne R (2019) Effect of media and fermentation conditions on surfactin and iturin homologues produced by Bacillus natto NT-6: LC–MS analysis. AMB Exp 9:1–9
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Upadhyay SK, Chauhan PK (2022) Optimization of eco-friendly amendments as sustainable asset for salt-tolerant plant growth-promoting bacteria mediated maize (Zea mays L.) plant growth Na uptake reduction and saline soil restoration. Environl Res. https://doi.org/10.1016/j.envres.2022.113081
Vargas L, Santa Brigida AB, Mota Filho JP, De Carvalho TG, Rojas CA, Vaneechoutte D, Van Bel M, Farrinelli L, Ferreira PC, Vandepoele K (2014) Drought tolerance conferred to sugarcane by association with Gluconacetobacter diazotrophicus: a transcriptomic view of hormone pathways. PLoS ONE 9:e114744
Wang D, Amornsiripanitch N, Dong X (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2:e123
Yan JY, Zhao WS, Chen Z, Xing QK, Zhang W, Chethana KT, Xue MF, Xu JP, Phillips AJ, Wang Y (2018) Comparative genome and transcriptome analyses reveal adaptations to opportunistic infections in woody plant degrading pathogens of Botryosphaeriaceae. DNA Res 25:87–102
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:1–12
Zander M, La Camera S, Lamotte O, Métraux JP, Gatz C (2010) Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J 61:200–210
Zavaliev R, Mohan R, Chen T, Dong X (2020) Formation of NPR1 condensates promotes cell survival during the plant immune response. Cell 182(1093–1108):e1018
Zhang X, Liu C-J (2015) Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol Plant 8:17–27
Zhang X, Wu F, Gu N, Yan X, Wang K, Dhanasekaran S, Gu X, Zhao L, Zhang H (2020) Postharvest biological control of Rhizopus rot and the mechanisms involved in induced disease resistance of peaches by Pichia membranefaciens. Postharvest Biol Technol 163:111146
Funding
This study was financially supported by the Research and Development Foundation of Yunnan Tobacco Company, China (Grant No. 2019530000241008).
Author information
Authors and Affiliations
Contributions
RJ, AA, PH, and SM contributed to conceptualization; RJ contributed to data curation; AA contributed to formal analysis; YC and YH contributed to funding acquisition; RJ, PH, and YC contributed to investigation; RJ, YW, JW, PH, GW, HY, JZ, and CL contributed to methodology; YW and YH contributed to project administration; , YW, CL, and YC contributed to resources; RJ, AA, and SM contributed to software; RJ, AA, and PH contributed to validation; YC and YH contributed to visualization; RJ and AA contributed to writing–original draft; and AA, SM, and YH contributed to writing–review & editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Handling Editor: Padmanabh Dwivedi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
344_2023_10922_MOESM1_ESM.docx
Supplementary file1 (DOCX 2499 KB)—Figure S1: Validation of RNA-seq results by qRT-PCR analysis. In total, 12 genes were randomly selected regulated in all treatment groups for real-time PCR analysis and alcohol dehydrogenase (ADH2) gene was used as internal reference. All the values are mean of three biological replicates; Table S1: Sequences of primers used in qRT-PCR for RNA-seq data validation
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiao, R., Ahmed, A., He, P. et al. Bacillus amyloliquefaciens Induces Resistance in Tobacco Against Powdery Mildew Pathogen Erysiphe cichoracearum. J Plant Growth Regul 42, 6636–6651 (2023). https://doi.org/10.1007/s00344-023-10922-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-023-10922-3