Abstract
Masson pine (Pinus massoniana) is primarily present in southwest of China, which are severely deficient in inorganic phosphate (Pi). Although some studies identified transcriptomic and proteomic responses to Pi deficiency in Masson pine seedlings, miRNAs and molecular responses in different tissues have not been well studied. To shed further light on the complex responses of Masson pine to Pi deficiency, a spatiotemporal experiment was performed to identify differentially expressed mRNAs and miRNAs under Pi deficiency. Spatiotemporal analyses of 72 RNA sequencing libraries provided a comprehensive overview of the dynamic responses of Masson pine to low-Pi stress. Differentially expressed gene analysis revealed several high-affinity phosphate transporter genes (PHT1-1, PHT1-4, PHT1-5 and PHT1-12) and a nitrate transporter gene (NRT), reflecting the crosstalk between nitrate and Pi homeostasis in plants. miRNA differential expression analysis identified several families that were associated with Pi deficiency, such as miR399. In addition, some other families were dramatically changed in response to Pi starvation, such as miR156, miR169, and some novel miRNAs. Integrated analysis of DE miRNAs and mRNAs indicated that “amino acid metabolism”, “energy metabolism” and “lipid metabolism” were most enriched under Pi deficiency. This study provided essential regulation information between miRNAs and target genes on the response of Masson pine seedlings to Pi deficiency, which will aid in further elucidation of the biological regulatory mechanisms of pines in response to low-Pi stress.








Similar content being viewed by others
Abbreviations
- P:
-
Phosphorus
- Pi:
-
Inorganic phosphate
- PHR1:
-
Phosphate starvation response1
- SPX:
-
SYG1/PHO81/XPR1
- PHO2:
-
Phosphate2
- PHTs:
-
Phosphate transporters
- PAPs:
-
Purple acid phosphatases
- DEGs:
-
Differentially expressed genes
- ERF:
-
Ethylene responsive factor
- PEPC:
-
Phosphoenolpyruvate carboxylase
- GDPDs:
-
Glycerophosphodiester phosphodiesterases
- SQD1:
-
UDP-sulfoquinovose synthase
- MGDs:
-
Monogalactosyldiacylglycerol synthases
- AP2 :
-
APETALA2
- RSA:
-
Root system architecture
- RPKM:
-
Reads per kilobase per million reads
- PCC:
-
Pearson correlation coefficient
- RT-qPCR:
-
Reverse transcription quantitative real-time polymerase chain reaction
References
Abel S (2017) Phosphate scouting by root tips. Curr Opin Plant Biol 39:168–177. https://doi.org/10.1016/j.pbi.2017.04.016
Abel S, Ticconi CA, Delatorre CA (2002) Phosphate sensing in higher plants. Physiol Plantarum 115:1–8. https://doi.org/10.1034/j.1399-3054.2002.1150101.x
Ames B (1966) Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol 8:115–118. https://doi.org/10.1016/0076-6879(66)08014-5
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Nat Prec 11:R106. https://doi.org/10.1038/npre.2010.4282.2
Andersson H, Bergström L, Djodjic F, Ulén B, Kirchmann H (2013) Topsoil and subsoil properties influence phosphorus leaching from four agricultural soils. J Environ Qual 42:455–463. https://doi.org/10.2134/jeq2012.0224
Anuradha M, Narayanan A (1991) Promotion of root elongation by phosphorus deficiency. Plant Soil 136:273–275. https://doi.org/10.1007/BF02150060
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29. https://doi.org/10.1038/75556
Bao H, Chen H, Chen M, Xu H, Huo X, Xu Q, Wang Y (2019) Transcriptome-wide identification and characterization of microRNAs responsive to phosphate starvation in Populus tomentosa. Funct Integr Genomics 19:953–972. https://doi.org/10.1007/s10142-019-00692-1
Bari R, Pant BD, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141:988–999. https://doi.org/10.1104/pp.106.079707
Chen ZC, Liao H (2016) Organic acid anions: an effective defensive weapon for plants against aluminum toxicity and phosphorus deficiency in acidic soils. J Genet Genomics 43:631–638. https://doi.org/10.1016/j.jgg.2016.11.003
Chen Y, Wu P, Zhao Q, Tang Y, Chen Y, Li M, Jiang H, Wu G (2018) Overexpression of a phosphate starvation response AP2/ERF gene from physic nut in Arabidopsis alters root morphological traits and phosphate starvation-induced anthocyanin accumulation. Front Plant Sci 9:1186. https://doi.org/10.3389/fpls.2018.01186
Cheng L, Bucciarelli B, Liu J, Zinn K, MillerS P-V, Allan D, Shen J, Vance CP (2011) White lupin cluster root acclimation to phosphorus deficiency and root hair development involve unique glycerophosphodiester phosphodiesterases. Plant Physiol 156(3):1131–1148
Chien PS, Chiang CB, Wang Z, Chiou TJ (2017) MicroRNA-mediated signaling and regulation of nutrient transport and utilization. Curr Opin Plant Biol 39:73–79. https://doi.org/10.1016/j.pbi.2017.06.007
Chiou TJ (2007) The role of microRNAs in sensing nutrient stress. Plant Cell Environ 30:323–332. https://doi.org/10.1111/j.1365-3040.2007.01643.x
Cui YN, Li XT, Yuan JZ, Wang FZ, Wang SM, Ma Q (2019) Nitrate transporter NPF7.3/NRT1.5 plays an essential role in regulating phosphate deficiency responses in Arabidopsis. Biochem Bioph Res Co 508:314–319. https://doi.org/10.1016/j.bbrc.2018.11.118
Daloso DM, dos Anjos L, Fernie AR (2016) Roles of sucrose in guard cell regulation. New Phytol 211:809–818. https://doi.org/10.1111/nph.13950
de Souza Campos PM, Cornejo P, Rial C, Borie F, Varela RM, Seguel A, López-Ráez JA (2019) Phosphate acquisition efficiency in wheat is related to root: shoot ratio, strigolactone levels, and PHO2 regulation. J Exp Bot 70:5631–5642. https://doi.org/10.1093/jxb/erz349
Duan K, Yi K, Dang L, Huang H, Wu W, Wu P (2008) Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 54:965–975. https://doi.org/10.1111/j.1365-313X.2008.03460.x
Duff SMG, Sarath G, Plaxton WC (1994) The role of acid-phosphatases in plant phosphorus-metabolism. Physiol Plantarum 4:791–800. https://doi.org/10.1111/j.1399-3054.1994.tb02539.x
Ernst J, Bar-Joseph Z (2006) STEM: a tool for the analysis of short time series gene expression data. BMC Bioinf 7:191. https://doi.org/10.1186/1471-2105-7-191
Fan F, Cui B, Zhang T, Qiao G, Ding G, Wen X (2014) The temporal transcriptomic response of Pinus massoniana seedlings to phosphorus deificiency. PLoS ONE 9:e105068. https://doi.org/10.1371/journal.pone.0105068
Fan F, Ding G, Wen X (2016) Proteomic analyses provide new insights into the response of Pinus massoniana seedlings to phosphorus deficiency. Proteomics 16:504–515. https://doi.org/10.1002/pmic.201500140
Fang Z, Shao C, Meng Y, Wu P, Chen M (2009) Phosphate signaling in Arabidopsis and Oryza sativa. Plant Sci 176:170–180. https://doi.org/10.1016/j.plantsci.2008.09.007
Foti M, Dho S, Fusconi A (2016) Root plasticity of Nicotiana tabacum in response to phosphorus starvation. Plant Biosystems-an International Journal Dealing with All Aspects of Plant Biology 150:429–435. https://doi.org/10.1080/11263504.2014.986245
Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15:2038–2043. https://doi.org/10.1016/j.cub.2005.10.016
Gho YS, An G, Park HM, Jung KH (2018) A systemic view of phosphate starvation-responsive genes in rice roots to enhance phosphate use efficiency in rice. Plant Biot Rep 12:249–264. https://doi.org/10.1007/s11816-018-0490-y
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhurry R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotech 29:644–652. https://doi.org/10.1038/nbt.1883
Hammond JP, Bennett MJ, Bowen HC, Broadley MR (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132:578–596. https://doi.org/10.1104/pp.103.020941
Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151:2120–2132. https://doi.org/10.1104/pp.109.147280
Hu B, Jiang Z, Wang W, Qiu Y, Zhang Z, Liu Y, Li A, Gao X, Liu L, Qian Y, Huang X, Yu F, Kang S, Wang Y, Xie J, Cao S, Zhang L, Wang Y, Xie Q, Kopriva S, Chu C (2019) Nitrate-NRT1.1B-SPX4 cascade integrates nitrogen and phosphorus signaling networks in plants. Nat Plants 5:401–413. https://doi.org/10.1038/s41477-019-0384-1
Huen A, Bally J, Smith P (2018) Identification and characterisation of microRNAs and their target genes in phosphate-starved Nicotiana benthamiana by small RNA deep sequencing and 5’RACE analysis. BMC Genomics 19:940. https://doi.org/10.1186/s12864-018-5258-9
Jain A, Nagarajan VK, Raghothama KG (2012) Transcriptional regulation of phosphate acquisition by higher plants. Cell Mol Life Sci 69:3207–3224. https://doi.org/10.1007/s00018-012-1090-6
Jez JM, Lee SG, Sherp AM (2016) The next green movement: plant biology for the environment and sustainability. Science 353:1241–1244. https://doi.org/10.1126/science.aag1698
Kalvari I, Nawrocki EP, Argasinska J, Quinones-Olvera N, Finn RD, Bateman A, Petrov AI (2018) Non-coding RNA analysis using the Rfam database. Curr Protoc Bioinformatics 62:e51. https://doi.org/10.1002/cpbi.51
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484. https://doi.org/10.1093/nar/gkm882
Kobayashi K, Awai K, Nakamura M, Nagatani A, Masuda T, Ohta H (2009) Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J 57:322–331. https://doi.org/10.1111/j.1365-313X.2008.03692.x
Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47:D155–D162. https://doi.org/10.1093/nar/gky1141
Kuo HF, Chiou TJ (2011) The role of microRNAs in phosphorus deficiency signaling. Plant Physiol 156:1016–1024. https://doi.org/10.1104/pp.111.175265
Lan P, Li W, Schmidt W (2012) Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol Cell Proteomics 11:1156–1166. https://doi.org/10.1074/mcp.M112.020461
Lapis-Gaza HR, Jost R, Finnegan PM (2014) Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1; 8 and PHT1; 9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol 14:334. https://doi.org/10.1186/s12870-014-0334-z
Li X, Zhang Y, Yin L, Lu J (2017) Overexpression of pathogen-induced grapevine TIR-NB-LRR gene VaRGA1 enhances disease resistance and drought and salt tolerance in Nicotiana benthamiana. Protoplasma 254:957–969. https://doi.org/10.1007/s00709-016-1005-8
Liu TY, Huang TK, Yang SY, Hong YT, Huang SM, Wang FN, Chiang SF, Tsai SY, Lu WC, Chiou TJ (2016) Identification of plant vacuolar transporters mediating phosphate storage. Nat Commun 7:11095. https://doi.org/10.1038/ncomms11095
Livak KJ, Schmittgen TD (2011) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ Ct method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Luo J, Liu Y, Zhang H, Wang J, Chen Z, Luo L, Liu G, Liu P (2020) Metabolic alterations provide insights into Stylosanthes roots responding to phosphorus deficiency. BMC Plant Biol. https://doi.org/10.1186/s12870-020-2283-z
Lv Q, Zhong Y, Wang Y, Wang Z, Zhang L, Shi J, Wu Z, Liu Y, Mao C, Yi K, Wu P (2014) SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26:1586–1597. https://doi.org/10.1105/tpc.114.123208
Mehra P, Giri J (2016) Rice and chickpea GDPDs are preferentially influenced by low phosphate and CaGDPD1 encodes an active glycerophosphodiester phosphodiesterase enzyme. Plant Cell Rep 35:1699–1717. https://doi.org/10.1007/s00299-016-1984-0
Mehra P, Pandey BK, Giri J (2017) Improvement in phosphate acquisition and utilization by a secretory purple acid phosphatase (OsPAP21b) in rice. Plant Biotechnol J 15:1054–1067. https://doi.org/10.1111/pbi.12699
Mehra P, Pandey BK, Verma L, Giri J (2019) A novel glycerophosphodiester phosphodiesterase improves phosphate deficiency tolerance in rice. Plant Cell Environ 42:1167–1179. https://doi.org/10.1111/pce.13459
Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD (2019) PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47:D419–D426. https://doi.org/10.1093/nar/gky1038
Miao J, Sun J, Liu D, Li B, Zhang A, Li Z, Tong Y (2009) Characterization of the promoter of phosphate transporter TaPHT1.2 differentially expressed in wheat varieties. J Genet Genomics 36:455–466. https://doi.org/10.1016/S1673-8527(08)60135-6
Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. https://doi.org/10.1038/nmeth.1226
Muchhal US, Pardo JM, Raghothama K (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93:10519–10523. https://doi.org/10.1073/pnas.93.19.10519
Nakamura Y (2013) Phosphate starvation and membrane lipid remodeling in seed plants. Prog Lipid Res 52:43–50. https://doi.org/10.1016/j.plipres.2012.07.002
Nakamura M, Köhler C, Hennig L (2019) Tissue-specific transposon-associated small RNAs in the gymnosperm tree. Norway Spruce BMC Genomics 20:997. https://doi.org/10.1186/s12864-019-6385-7
Naumann C, Müller J, Sakhonwasee S, Wieghaus A, Hause G, Heisters M, Bürstenbinder K, Abel S (2019) The local phosphate deficiency response activates endoplasmic reticulum stress-dependent autophagy. Plant Physiol 179:460–476. https://doi.org/10.1104/pp.18.01379
Niu SH, Liu C, Yuan HW, Li P, Li Y, Li W (2015) Identification and expression profiles of sRNAs and their biogenesis and action-related genes in male and female cones of Pinus tabuliformis. BMC Genomics 16:693. https://doi.org/10.1186/s12864-015-1885-6
O′Rourke JA, Yang SS, Miller SS, Bucciarelli B, Liu J, Rydeen A, Bozsoki Z, Uhde-Stone C, Tu ZJ, Allan D, Gronwald JW, Vance CP (2013) An RNASeq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol 161:705–724. https://doi.org/10.1104/pp.112.209254
Osorio MB, Ng S, Berkowitz O, De Clercq I, Mao C, Shou H, Whelan J, Jost R (2019) SPX4 acts on PHR1-dependent and independent regulation of shoot phosphorus status in Arabidopsis. Plant Physiol 181:332–352. https://doi.org/10.1104/pp.18.00594
Ouyang X, Hong X, Zhao X, Zhang W, He X, Ma W, Teng W, Tong Y (2016) Knock out of the PHOSPHATE 2 gene TaPHO2-A1 improves phosphorus uptake and grain yield under low phosphorus conditions in common wheat. Sci Rep 6:29850. https://doi.org/10.1038/srep29850
Park MR, Baek SH, De los Reyes BG, Yun SJ, Hasenstein KH (2012) Transcriptome profiling characterizes phosphate deficiency effects on carbohydrate metabolism in rice leaves. J Plant Physiol 169:193–205. https://doi.org/10.1016/j.jplph.2011.09.002
Park BS, Seo JS, Chua NH (2014) Nitrogen limitation adaptation recruits Phosphate2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26:454–464. https://doi.org/10.1105/tpc.113.120311
Peñaloza E, Muñoz G, Salvo-Garrido H, Silva H, Corcuera LJ (2005) Phosphate deficiency regulates phosphoenolpyruvate carboxylase expression in proteoid root clusters of white lupin. J Exp Bot 56:145–153. https://doi.org/10.1093/jxb/eri008
Qin L, Zhao J, Tian J, Chen L, Sun Z, Guo Y, Lu X, Gu M, Xu G, Liao H (2012) The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol 159:1634–1643. https://doi.org/10.1104/pp.112.199786
Ren Y, Sun F, Hou J, Chen L, Zhang Y, Kang X, Wang Y (2015) Differential profiling analysis of miRNAs reveals a regulatory role in low N stress response of Populus. Funct Integr Genomics 15:93–105. https://doi.org/10.1007/s10142-014-0408-x
Ren P, Meng Y, Li B, Ma X, Si E, Lai Y, Wang J, Yao L, Yang K, Shang X, Wang H (2018) Molecular mechanisms of acclimatization to phosphorus starvation and recovery underlying full-length transcriptome profiling in barley (Hordeum vulgare L.). Front Plant Sci 9:500. https://doi.org/10.3389/fpls.2018.00500
Rodrigues AS, Chaves I, Costa BV, Lin YC, Lopes S, Milhinhos A, Van de Peer Y, Miguel CM (2019) Small RNA profiling in Pinus pinaster reveals the transcriptome of developing seeds and highlights differences between zygotic and somatic embryos. Sci Rep 9:11327. https://doi.org/10.1038/s41598-019-47789-y
Ruan W, Guo M, Wang X, Guo Z, Xu Z, Xu L, Zhao H, Sun H, Yan C, Yi K (2019) Two RING-finger ubiquitin E3 ligases regulate the degradation of SPX4, an internal phosphate sensor, for phosphate homeostasis and signaling in rice. Mol Plant 12:1060–1074. https://doi.org/10.1016/j.molp.2019.04.003
Sakuraba Y, Kanno S, Mabuchi A, Monda K, Iba K, Yanagisawa S (2018) A phytochrome-B-mediated regulatory mechanism of phosphorus acquisition. Nat Plants 4:1089–1101. https://doi.org/10.1038/s41477-018-0294-7
Schroeder JI, Delhaize E, Frommer WB, Guerinot ML, Harrison MJ, Herrera-Estrella L, Horie T, Kochian LV, Munns R, Nishizawa NK, Tsay Y, Sanders D (2013) Using membrane transporters to improve crops for sustainable food production. Nature 497:60–66. https://doi.org/10.1038/nature11909
Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J (2012) The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol 193:842–851. https://doi.org/10.1111/j.1469-8137.2011.04002.x
Secco D, Jabnoune M, Walker H, Shou H, Wu P, Poirier Y, Whelan J (2013) Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25:4285–4303. https://doi.org/10.1105/tpc.113.117325
Shemi A, Schatz D, Fredricks HF, Van Mooy BA, Porat Z, Vardi A (2016) Phosphorus starvation induces membrane remodeling and recycling in Emiliania huxleyi. New Phytol 211:886–898. https://doi.org/10.1111/nph.13940
Shen J, Yan L, Zhang J, Li H, Bai Z, Chen X, Zhang W, Zhang F (2011) Phosphorus dynamics: from soil to plant. Plant Physiol 156:997–1005. https://doi.org/10.1104/pp.111.175232
Shin H, Shin HS, Dewbre GR, Harrison MJ (2004) Phosphate transport in Arabidopsis: Pht1; 1 and Pht1; 4 play a major role in phosphate acquisition from both low- and high- phosphate environments. Plant J 39:629–642. https://doi.org/10.1111/j.1365-313X.2004.02161.x
Song H, Cai Z, Liao J, Tang D, Zhang S (2019) Sexually differential gene expressions in poplar roots in response to nitrogen deficiency. Tree Physiol 39:1614–1629. https://doi.org/10.1093/treephys/tpz057
Strock CF, De La Riva LM, Lynch JP (2018) Reduction in root secondary growth as a strategy for phosphorus acquisition. Plant Physiol 176:691–703. https://doi.org/10.1104/pp.17.01583
Tawaraya K, Honda S, Cheng W, Chuba M, Okazaki Y, Saito K, Oikawa A, Maruyama H, Wasaki J, Wagatsuma T (2018) Ancient rice cultivar extensively replaces phospholipids with non-phosphorus glycolipid under phosphorus deficiency. Physiol Plantarum 163:297–305. https://doi.org/10.1111/ppl.12699
Tong Y, Zhang W, Wang X, Couture RM, Larssen T, Zhao Y, Li J, Liang H, Liu X, Bu X, He W, Zhang Q, Lin Y (2006) Decline in Chinese lake phosphorus concentration accompanied by shift in source since 2006. Nat Geosci 10:507–511. https://doi.org/10.1038/ngeo2967
Trujillo-Hernandez JA, Bariat L, Enders TA, Strader LC, Reichheld JP, Belin C (2020) A glutathione- dependent control of the indole butyric acid pathway supports Arabidopsis root system adaptation to phosphate deprivation. J Exp Bot 16:4843–4857. https://doi.org/10.1093/jxb/eraa195
Vance CP, Uhde-Stone C, Allan DLP (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157:423–447. https://doi.org/10.1046/j.1469-8137.2003.00695.x
Veneklaas EJ, Lambers H, Bragg J, Finnegan PM, Lovelock CE, Plaxton WC, Price CA, Scheible WR, Shane MW, White PJ, Raven JA (2012) Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol 195:306–320. https://doi.org/10.1111/j.1469-8137.2012.04190.x
Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138. https://doi.org/10.1093/bioinformatics/btp612
Wild R, Gerasimaite R, Jung JY, Truffault V, Pavlovic I, Schmidt A, Saiardi A, Jessen HJ, Poirier Y, Hothorn M, Mayer A (2016) Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352:986–990. https://doi.org/10.1126/science.aad9858
Xie R, Zhang J, Ma Y, Pan X, Dong C, Pang S, He S, Deng L, Yi S, Zheng Y, Lv Q (2017) Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots. Sci Rep 7:42094. https://doi.org/10.1038/srep42094
Yan T, Yoo D, Berardini TZ, Mueller LA, Weems DC, Weng S, Cherry JM, Rhee SY (2005) PatMatch: a program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res 33:W262–W266. https://doi.org/10.1093/nar/gki368
Zhang Y, Zhou Z, Ma X, Jin G (2010) Foraging ability and growth performance of four subtropical tree species in response to heterogeneous nutrient environments. J Res 15:91–98. https://doi.org/10.1007/s10310-009-0153-5
Zhang Z, Liao H, Lucas WJ (2014) Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J Integr Plant Biol 56:192–220. https://doi.org/10.1111/jipb.12163
Zhang Z, Wang Z, Wang X, Liu D (2019) Blue light-triggered chemical reactions underlie phosphate deficiency-induced inhibition of root elongation of Arabidopsis seedlings grown in Petri dishes. Mol Plant 11:1515–1523. https://doi.org/10.1016/j.molp.2019.08.001
Zhao M, Ding H, Zhu JK, Zhang FS, Li WX (2011) Involvement of miR169 in the nitrogen-starvation response in Arabidopsis. New Phytol 190:906–915. https://doi.org/10.1111/j.1469-8137.2011.03647.x
Zhao P, Wang L, Yin H (2018) Transcriptional responses to phosphate starvation in Brachypodium distachyon roots. Plant Physiol Bioch 122:113–120. https://doi.org/10.1016/j.plaphy.2017.11.010
Funding
This study was funded by the National Key Research and Development Project, P. R. China (2017YFD0600302); National Natural Science Foundation of China (31660185); Science and Technology of Guizhou Province, P. R. China (20175788); Innovation Driven Development Special Fund of Guangxi, P. R. China (AA17204087-4); and the Post-National Key Research and Development Project, P. R. China (20185261).
Author information
Authors and Affiliations
Contributions
FF conceived, designed the experiments and wrote the paper. XS performed experiments. GD contributed reagents and materials, revised manuscript. ZZ performed experiments. JT collected data. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Handling Editor: Mikihisa Umehara.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
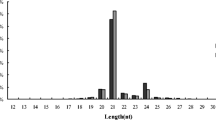
Length distribution of transcripts using next-generation sequencing. (A)Length distribution of Masson pine unigenes using next-generation sequencing. (B) miRNA length distribution of Masson pine.
Figure S2
Venn diagram of the unigenes annotated by four databases (KEGG, KOG, Nr and Swissprot).
Appendix A: Supplementary Data
Appendix A: Supplementary Data
Illumina reads of all samples have been submitted to the Sequence Read Archive at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/sra) under Accession Number PRJNA641031.
Rights and permissions
About this article
Cite this article
Fan, F., Shang, X., Ding, G. et al. Integrated mRNA and miRNA Expression Analyses of Pinus massoniana Roots and Shoots in Long-Term Response to Phosphate Deficiency. J Plant Growth Regul 41, 2949–2966 (2022). https://doi.org/10.1007/s00344-021-10486-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-021-10486-0