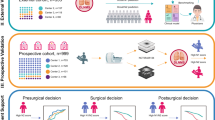
Abstract
Objectives
To develop and validate CT-based deep learning (DL) models that learn morphological and histopathological features for lung adenocarcinoma prognostication, and to compare them with a previously developed DL discrete-time survival model.
Methods
DL models were trained to simultaneously predict five morphological and histopathological features using preoperative chest CT scans from patients with resected lung adenocarcinomas. The DL score was validated in temporal and external test sets, with freedom from recurrence (FFR) and overall survival (OS) as outcomes. Discrimination was evaluated using the time-dependent area under the receiver operating characteristic curve (TD-AUC) and compared with the DL discrete-time survival model. Additionally, we performed multivariable Cox regression analysis.
Results
In the temporal test set (640 patients; median age, 64 years), the TD-AUC was 0.79 for 5-year FFR and 0.73 for 5-year OS. In the external test set (846 patients; median age, 65 years), the TD-AUC was 0.71 for 5-year OS, equivalent to the pathologic stage (0.71 vs. 0.71 [p = 0.74]). The prognostic value of the DL score was independent of clinical factors (adjusted per-percentage hazard ratio for FFR (temporal test), 1.02 [95% CI: 1.01–1.03; p < 0.001]; OS (temporal test), 1.01 [95% CI: 1.002–1.02; p = 0.01]; OS (external test), 1.01 [95% CI: 1.005–1.02; p < 0.001]). Our model showed a higher TD-AUC than the DL discrete-time survival model, but without statistical significance (2.5-year OS: 0.73 vs. 0.68; p = 0.13).
Conclusion
The CT-based prognostic score from collective deep learning of morphological and histopathological features showed potential in predicting survival in lung adenocarcinomas.
Clinical relevance statement
Collective CT-based deep learning of morphological and histopathological features presents potential for enhancing lung adenocarcinoma prognostication and optimizing pre-/postoperative management.
Key Points
• A CT-based prognostic model was developed using collective deep learning of morphological and histopathological features from preoperative CT scans of 3181 patients with resected lung adenocarcinoma.
• The prognostic performance of the model was comparable-to-higher performance than the pathologic T category or stage.
• Our approach yielded a higher discrimination performance than the direct survival prediction model, but without statistical significance (0.73 vs. 0.68; p=0.13).






Similar content being viewed by others
Abbreviations
- DL:
-
Deep learning
- FFR:
-
Freedom from recurrence
- HR:
-
Hazard ratio
- IQR:
-
Interquartile range
- OS:
-
Overall survival
- ROC:
-
Receiver operating characteristic
- TD-AUC:
-
Time-dependent area under the ROC curve
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2022) Cancer statistics, 2022. CA Cancer J Clin 72:7–33
Cancer Research UK. Lung cancer survival statistics. Available via https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/lung-cancer/survival#heading-Three. Accessed 24 Jan 2022
Humphries SM, Notary AM, Centeno JP et al (2020) Deep learning enables automatic classification of emphysema pattern at CT. Radiology 294:434–444
Venkadesh KV, Setio AAA, Schreuder A et al (2021) Deep learning for malignancy risk estimation of pulmonary nodules detected at low-dose screening CT. Radiology 300:438–447
Oh AS, Baraghoshi D, Lynch DA et al (2022) Emphysema progression at CT by deep learning predicts functional impairment and mortality: results from the COPDGene Study. Radiology 304:672–679
Jiang Y, Zhang Z, Yuan Q et al (2022) Predicting peritoneal recurrence and disease-free survival from CT images in gastric cancer with multitask deep learning: a retrospective study. Lancet Digit Health 4:e340–e350
Zhong Y, She Y, Deng J et al (2022) Deep learning for prediction of N2 metastasis and survival for clinical stage I non-small cell lung cancer. Radiology 302:200–211
Torres FS, Akbar S, Raman S et al (2021) End-to-end non-small-cell lung cancer prognostication using deep learning applied to pretreatment computed tomography. JCO Clin Cancer Inform 5:1141–1150
Hosny A, Parmar C, Coroller TP et al (2018) Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med 15:e1002711
Huang B, Sollee J, Luo YH et al (2022) Prediction of lung malignancy progression and survival with machine learning based on pre-treatment FDG-PET/CT. EBioMedicine 82:104127
Kim H, Goo JM, Lee KH, Kim YT, Park CM (2020) Preoperative CT-based deep learning model for predicting disease-free survival in patients with lung adenocarcinomas. Radiology 296:216–224
Clark TG, Bradburn MJ, Love SB, Altman DG (2003) Survival analysis part I: basic concepts and first analyses. Br J Cancer 89:232–238
Hattori A, Hirayama S, Matsunaga T et al (2019) Distinct clinicopathologic characteristics and prognosis based on the presence of ground glass opacity component in clinical stage IA lung adenocarcinoma. J Thorac Oncol 14:265–275
Kawase A, Yoshida J, Miyaoka E et al (2013) Visceral pleural invasion classification in non-small-cell lung cancer in the 7th edition of the tumor, node, metastasis classification for lung cancer: validation analysis based on a large-scale nationwide database. J Thorac Oncol 8:606–611
Yim J, Zhu LC, Chiriboga L, Watson HN, Goldberg JD, Moreira AL (2007) Histologic features are important prognostic indicators in early stages lung adenocarcinomas. Mod Pathol 20:233–241
Russell PA, Wainer Z, Wright GM, Daniels M, Conron M, Williams RA (2011) Does lung adenocarcinoma subtype predict patient survival?: a clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary lung adenocarcinoma classification. J Thorac Oncol 6:1496–1504
Shimada Y, Saji H, Yoshida K et al (2012) Pathological vascular invasion and tumor differentiation predict cancer recurrence in stage IA non-small-cell lung cancer after complete surgical resection. J Thorac Oncol 7:1263–1270
Detterbeck FC, Boffa DJ, Kim AW, Tanoue LT (2017) The eighth edition lung cancer stage classification. Chest 151:193–203
Lee KH, Lee JH, Park S et al (2023) Computed tomography–based prognostication in lung adenocarcinomas through histopathological feature learning: a retrospective multicenter study. Ann Am Thorac Soc 20:1020–1028. https://doi.org/10.1513/AnnalsATS.202210-895OC
Lim WH, Lee KH, Lee JH et al (2023) Diagnostic performance and prognostic value of CT-defined visceral pleural invasion in early-stage lung adenocarcinomas. Eur Radiol. https://doi.org/10.1007/s00330-023-10204-2
Soffer S, Ben-Cohen A, Shimon O, Amitai MM, Greenspan H, Klang E (2019) Convolutional neural networks for radiologic images: a radiologist’s guide. Radiology 290:590–606
Hong S, Won YJ, Lee JJ et al (2021) Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2018. Cancer Res Treat 53:301–315
Blanche P, Dartigues JF, Jacqmin-Gadda H (2013) Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med 32:5381–5397
Uno H, Cai T, Pencina MJ, D’Agostino RB, Wei LJ (2011) On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med 30:1105–1117
Kim H, Lee JH, Kim HJ, Park CM, Wu HG, Goo JM (2021) Extended application of a CT-based artificial intelligence prognostication model in patients with primary lung cancer undergoing stereotactic ablative radiotherapy. Radiother Oncol 165:166–173
Singh T, Ghosh A, Khandelwal N (2017) Dimensional reduction and feature selection: principal component analysis for data mining. Radiology 285:1055–1056
Heagerty PJ, Lumley T, Pepe MS (2000) Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 56:337–344
Choi Y, Aum J, Lee SH et al (2021) Deep learning analysis of CT images reveals high-grade pathological features to predict survival in lung adenocarcinoma. Cancers (Basel) 13
Funding
This study was supported by the Seoul National University Hospital Research Fund (grant number: 04-2020-2040 and 03-2022-2170) and by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. RS-2023-00207978). However, the funders had no role in the study design; in the collection, analysis, and interpretation of the data; in the writing of the report; and in the decision to submit the article for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Hyungjin Kim.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors (Hyungjin Kim) has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Study subjects or cohorts overlap
Some patients in our datasets have been reported previously (Lee et al [19]; 2717/3181); Lim et al [20]; 681/3181)). However, none of the prior studies dealt with the multitask learning of both morphological and histopathological features for prognostication.
Methodology
• retrospective
• diagnostic or prognostic study
• multicenter study
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lee, T., Lee, K.H., Lee, J.H. et al. Prognostication of lung adenocarcinomas using CT-based deep learning of morphological and histopathological features: a retrospective dual-institutional study. Eur Radiol (2023). https://doi.org/10.1007/s00330-023-10306-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00330-023-10306-x