Abstract
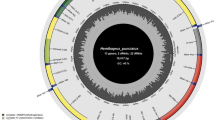
Mitochondrial DNA (mtDNA) has been widely used in species identification and genetic diversification. Comparisons among mtDNAs of closely related species are valuable for phylogenetic studies. However, only the partial mtDNA Cytb gene and the D-loop sequences were used to analysis the phylogenetic relationship between donkey breeds due to lack of complete mitochondrial genome. Dezhou donkey, as a bigger somatotype ass, is one of Chinese domestic donkey breeds, and used by many places as breeding stock. To further investigate the phylogenetic relationship of Dezhou donkey with other breeds, the complete mtDNA was firstly sequenced and de novo assembled using next generation sequence data from total genomic DNA. The genome was 16,813 bp in length (NCBI submission number: KT182635) and contained 13 protein coding genes, 2 ribosomal RNA genes, 25 transfer RNA genes, and 1 control region. Based on the novel complete mtDNA sequence, the sequences of 13 protein coding genes and 2 ribosomal RNA genes were amplifying in other 2 Dezhou donkeys and 3 Yunnan donkeys, respectively. The pattern of genetic variation in horse, wild ass and domestic donkeys among these 15 genes indicated the sequence polymorphisms. The more accurate phylogenetic relationships of donkey species (Dezhou donkey, Yunnan donkey and previously published donkeys) were first obtained using the combined sequences of 12S rRNA+16S rRNA+13 protein-coding genes. Molecular-based phylogeny supported the hypothesis that Chinese domestic donkey breeds may have originated from Somali wild ass, not from Asian wild ass by analyzing mitochondrial genomes.



Similar content being viewed by others
References
Achilli A, Olivier A, Soares P, Lancioni H et al (2012) Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc Natl Acad Sci 109:2449–2454
Chen S, Zhou F, Xiao H, Sha T, Wu S, Zhang Y (2006) Mitochondrial DNA diversity and population structure of four Chinese donkey breeds. Anim Genet 37:422–431
Hegedusova E, Brejova B, Tomaska L, Sipiczki M, Nosek J (2014) Mitochondrial genome of the basidiomycetous yeast Jaminaea angkorensis. Curr Genet 60:49–59. doi:10.1007/s00294-013-0410
Iorizzo M, Senalik D, Szklarczyk M, Grzebelus D, Spooner D, Simon P (2012) De novo assembly of the carrot mitochondrial genome using next generation sequencing of whole genomic DNA provides first evidence of DNA transfer into an angiosperm plastid genome. BMC Plant Biol 12:61
Jonsson H, Schubert M, Orlando A, Ginolhac A, Petersen L et al (2014) Speciation with gene flow in equids despite extensive chromosomal plasticity. Proc Natl Acad Sci 111:18655–18660
Kumeta Y, Maruyama T, Asama H, Yamamoto Y, Hakamatsuka T, Goda Y (2014) Species identification of Asini Corii Collas (donkey glue) by PCR amplification of cytochrome b gene. J Nat Med 68:181–185
Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Yang H, Wang J, Wang J (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272
Lippold S, Matzke J, Reissmann M, Hofreiter M (2011) Whole mitochondrial genome sequencing of domestic horses reveals incorporation of extensive wild horse diversity during domestication. BMC Evol Biol 11:328
Liu F, Pang SJ (2015) Mitochondrial genome of Turbinaria ornate (Sargassaceae, Phaeophyceae): comparative mitogenomics of brown algae. Curr Genet. doi:10.1007/s00294-015-0488-8
Lohse M, Drechsel O, Kahlau S, Bock R (2013) Organellar Genome DRAW- a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res 41:575–581
Mendez J, Pereira A, Avellanet R, Dzama K, Jordana J (2004) Mitochondrial DNA variation and genetic relationships in Spanish donkey breeds (Equus asinus). J Anim Breed Genet 121:319–330
Mindell D, Sorenson M, Dimcheff D (1998) An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Mol Biol Evol 15:1568–1571
Oakenfull E, Lim H, Ryder O (2000) A survey of equid mitochondrial DNA: implication for the evolution, genetic diversity and conservation of Equus. Conserv Genet 1:341–355
Pereira A, England P, Ferrand N, Jordan S, Bakhiet A, Abdalla M et al (2004) African origins of the domestic donkey. Science 304:1781
Smith D, Pearson R (2005) A review of the factors affecting the survival of donkeys in semi-arid regions of Sub-Saharan Africa. Trop Anim Health Pro 37:1–19
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5:molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tanaka Y, Tsuda M, Yasumoto K, Terachi T, Yamagishi H (2014) The complete mitochondrial genome sequence of Brassica oleracea and analysis of coexisting mitotypes. Curr Genet 60:277–284. doi:10.1007/s00294-014-0433-2
Valach M, Pryszcz L, Tomaska L, Gacser A, Gabaldon T, Nosek J (2012) Mitochondrial genome variability within the Candida parapsilosis species complex. Mitochondrion 12:514–519
Vilstrup J, Orlando A, Stiller M, Ginolhac A, Raghavan M, Nielsen S, Weinstock J et al (2013) Mitochondrial phylogenomics of modern and ancient equids. PLoS ONE 8:e55950
Wilson R (1990) The donkey. In: Payne WJA (ed) An introduction to animal husbandry in the tropics, pp 581–603
Wyman S, Jansen R, Boore J (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255
Xie C (1987) Horse and Ass Breeds in China. Shanghai Science and Technology Press, Shanghai
Xu XF, Arnason U (1994) The complete mitochondrial DNA sequence of the horse, Equus caballus: extensive heteroplasmy of the control region. Gene 148:357–362
Xu X, Gullberg A, Arnason U (1996) The complete mitochondrial DNA (mtDNA) of the donkey and mtDNA comparisons among four closely related mammalian species-pairs. J Mol Evol 43:438–446
Zhang BW (2011) China National Commission of Animal Genetic Resources. Animal genetic resources in China: Horses, Donkeys, Camels. China Agricultural Press, China
Zhang Y, Xie Z, Xie L, Tan W, Liu J, Deng X, Xie Z et al (2014) Analysis of the Rongshui Xiang duck (Anseriformes, Anatidae, Anas) mitochondrial DNA. Mitochondrial DNA. doi:10.3109/19401736.2014.971263
Zhao L, Liu GH, Zhao GH, Cai JZ, Zhu XQ, Qian AD (2015) Genetic differences between Chaberti aovina and C. erschowi revealed by sequence analysis of four mitochondrial genes. Mitochondrial DNA. doi:10.3109/19401736.2013.843089
Acknowledgments
This study was supported by Natural Science Foundation of Shandong Province (Grant No. ZR2015PC010), the Agriculture Major Innovation Project in Shandong province, Well-bred Program of Shandong Province (Grant no. 2014LZ), and Joint Innovation Funds of Dong E E Jiao and Shandong Academy of Agricultural Sciences. The authors alone are responsible for the content and writing of the paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Y. Sun and Q. Jiang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, Y., Jiang, Q., Yang, C. et al. Characterization of complete mitochondrial genome of Dezhou donkey (Equus asinus) and evolutionary analysis. Curr Genet 62, 383–390 (2016). https://doi.org/10.1007/s00294-015-0531-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-015-0531-9