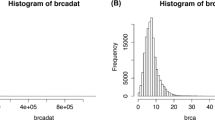
Abstract
Data taking values on discrete sample spaces are the embodiment of modern biological research. “Omics” experiments based on high-throughput sequencing produce millions of symbolic outcomes in the form of reads (i.e., DNA sequences of a few dozens to a few hundred nucleotides). Unfortunately, these intrinsically non-numerical datasets often deviate dramatically from natural assumptions a practitioner might make, and the possible sources of this deviation are usually poorly characterized. This contrasts with numerical datasets where Gaussian-type errors are often well-justified. To overcome this hurdle, we introduce the notion of latent weight, which measures the largest expected fraction of samples from a probabilistic source that conform to a model in a class of idealized models. We examine various properties of latent weights, which we specialize to the class of exchangeable probability distributions. As proof of concept, we analyze DNA methylation data from the 22 human autosome pairs. Contrary to what is usually assumed in the literature, we provide strong evidence that highly specific methylation patterns are overrepresented at some genomic locations when latent weights are taken into account.






Similar content being viewed by others
Notes
In other words, in this example, \(\mathcal {Q}\) denotes the set of product measures of the form \((\mu \otimes \nu )\), with \(\mu \) and \(\nu \) probability models supported on \(\{0,1\}\).
References
Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE (2012) methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 13(10):R87
Arellano-Valle RB, Genton MG (2008) On the exact distribution of the maximum of absolutely continuous dependent random variables. Stat Probab Lett 78(1):27–35
Bernstein B, Birney E, Dunham I, Green E, Gunter C, Snyder M, ENCODE Project Consortium, Hubbard T (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489(7414):57–74
Bickel PJ, Freedman DA (1981) Some asymptotic theory for the bootstrap. Ann Stat 9(6):1196–1217
Chestnut S, Lladser ME (2010) Occupancy distributions via Doeblin’s ergodicity coefficient. In: Proceedings of discrete mathematics and theoretical computer science, vol AM, pp 79–92
Core LJ, Waterfall JJ, Lis JT (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322(5909):1845–1848
Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, Onate KC, Graham K, Miyasato SR, Dreszer TR, Strattan JS, Jolanki O, Tanaka FY, Cherry JM (2017) The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 46(D1):D794–D801
De Finetti B (1937) La prévision: ses lois logiques, ses sources subjectives. Annales de l’institut Henri Poincaré 7(1):1–68
Diaconis P (1977) Finite forms of de Finetti’s theorem on exchangeability. Synthese 36(2):271–281
Diaconis P, Freedman D (1980) Finite exchangeable sequences. Ann Probab 8(4):745–764
Gnedin AV (1996) A class of exchangeable sequences. Stat Probab Lett 28(2):159–164
Good PI (2002) Extensions of the concept of exchangeability and their applications. J Mod Appl Stat Methods 1(2):34
Hall P, Härdle W, Simar L (1993) On the inconsistency of bootstrap distribution estimators. CORE Discussion Papers RP 1062, Université catholique de Louvain, Center for Operations Research and Econometrics (CORE). https://EconPapers.repec.org/RePEc:cor:louvrp:1062
Hampton J, Lladser ME (2012) Estimation of distribution overlap of urn models. PLoS ONE 7(11):e42368
Hansen KD, Langmead B, Irizarry RA (2012) BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol 13(10):R83
Huber PJ (1964) Robust estimation of a location parameter. Ann Math Stat 35(1):73–101
Huber PJ (1965) A robust version of the probability ratio test. Ann Math Stat 36(6):1753–1758
Ilie L, Fazayeli F, Ilie S (2010) HiTEC: accurate error correction in high-throughput sequencing data. Bioinformatics 27(3):295–302
Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13(1):484–492
Jones PL, Veenstra GCJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP (1998) Methylated dna and mecp2 recruit histone deacetylase to repress transcription. Nat Genet 19(2):187
Kingman JFC (1982) On the genealogy of large populations. J Appl Probab 19(A):27–43
Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27(11):1571–1572
Lindvall T (1992) Lectures on the coupling method. Wiley series in probability and statistics—applied probability and statistics section. Wiley
Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in arabidopsis. Cell 133(3):523–536
Lladser ME, Azofeifa JG, Allen MA, Dowell RD (2017) RNA Pol II transcription model and interpretation of GRO-seq data. J Math Biol 74(1–2):77–97
Lladser ME, Chestnut S (2013) Approximation of sojourn-times via maximal couplings: motif frequency distributions. J Math Biol 69
Lladser ME, Goeuet R, Reeder J (2011) Extrapolation of urn models via poissonization: accurate measurements of the microbial unknown. PLoS One 6(6)
Lou DI, Hussmann JA, McBee RM, Acevedo A, Andino R, Press WH, Sawyer SL (2013) High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc Natl Acad Sci 110(49):19872–19877. https://doi.org/10.1073/pnas.1319590110
Medvedev P, Scott E, Kakaradov B, Pevzner P (2011) Error correction of high-throughput sequencing datasets with non-uniform coverage. Bioinformatics 27(13):i137–i141
National Center for Biotechnology Information: Contamination in Sequence Databases. https://www.ncbi.nlm.nih.gov/tools/vecscreen/contam/. Accessed: 01-2020
Newcomb S (1886) A generalized theory of the combination of observations so as to obtain the best result. Am J Math 8(4):343–366
Park PJ (2009) Chip-seq: advantages and challenges of a maturing technology. Nat Rev Genet 10(10):669
Pearson A, Lladser ME (2020) Hidden independence in unstructured probabilistic models. In: 31st International conference on probabilistic, combinatorial and asymptotic methods for the analysis of algorithms (AofA 2020), Leibniz International Proceedings in Informatics (LIPIcs), vol 159, pp 23:1–23:13. Schloss Dagstuhl–Leibniz-Zentrum für Informatik, Dagstuhl, Germany. https://doi.org/10.4230/LIPIcs.AofA.2020.23. https://drops.dagstuhl.de/opus/volltexte/2020/12053
Pearson A, Lladser ME (2021) Post-processed DNA methylation data. https://doi.org/10.6084/m9.figshare.16983499.v1
Posfai J, Roberts RJ (1992) Finding errors in DNA sequences. Proc Natl Acad Sci 89(10):4698–4702. https://doi.org/10.1073/pnas.89.10.4698
Punzo A, McNicholas PD (2016) Parsimonious mixtures of multivariate contaminated normal distributions. Biom J 58(6):1506–1537
Robertson KD (2005) DNA methylation and human disease. Nat Rev Genet 6(8):597
Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW (2014) Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12
Schmitt MW, Fox EJ, Salk JJ (2014) Risks of double-counting in deep sequencing. Proc Natl Acad Sci 111(16):E1560–E1560
Song Q, Decato B, Hong EE, Zhou M, Fang F, Qu J, Garvin T, Kessler M, Zhou J, Smith AD (2013) A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS ONE 8(12):e81148
Stinson LF, Keelan JA, Payne MS (2019) Identification and removal of contaminating microbial DNA from PCR reagents: impact on low-biomass microbiome analyses. Lett Appl Microbiol 68
Suzuki MM, Bird A (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9(6):465
Tukey JW (1960) A survey of sampling from contaminated distributions. Contrib Probab Stat (in: Olkin I et al., eds) pp 448–485
van der Vaart AW (1998) Asymptotic statistics. Cambridge Series in Statistical and Probabilistic Mathematics. Cambridge University Press
Wang Z, Gerstein M, Synder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10(1):57–63
Acknowledgements
We thank a reviewer for their thorough reading and constructive remarks about our manuscript.
Funding
This work was partially supported by National Science Foundation Graduate Research Fellowship Program Grant No. 2016198773 (Pearson), and National Science Foundation IGERT Grant No. 1144807.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pearson, A., Lladser, M.E. On latent idealized models in symbolic datasets: unveiling signals in noisy sequencing data. J. Math. Biol. 87, 26 (2023). https://doi.org/10.1007/s00285-023-01961-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00285-023-01961-1