Abstract
Under-reporting in epidemics, when it is ignored, leads to under-estimation of the infection rate and therefore of the reproduction number. In the case of stochastic models with temporal data, a usual approach for dealing with such issues is to apply data augmentation techniques through Bayesian methodology. Departing from earlier literature approaches implemented using reversible jump Markov chain Monte Carlo (RJMCMC) techniques, we make use of approximations to obtain faster estimation with simple MCMC. Comparisons among the methods developed here, and with the RJMCMC approach, are carried out and highlight that approximation-based methodology offers useful alternative inference tools for large epidemics, with a good trade-off between time cost and accuracy.




Similar content being viewed by others
References
Althaus CL (2014) Estimating the reproduction number of ebola virus (EBOV) during the 2014 outbreak in West Africa. PLoS Curr. Outbreaks. doi:10.1371/currents.outbreaks.91afb5e0f279e7f29e7056095255b288
Andersson H, Britton T (2000) Stochastic epidemic models and their statistical analysis. Springer, Berlin
Bjornstad ON, Finkenstadt BF, Grenfell BT (2002) Dynamics of measles epidemics: estimating scaling of transmission rates using a time series SIR model. Ecol Monogr 72(2):169–184
Chis-Ster I, Ferguson NM (2007) Transmission parameters of the 2001 foot and mouth epidemic in Great Britain. PLoS One 2(6):e502
Chowell G, Hengartner NW, Castillo-Chavez C, Fenimore PW, Hyman JM (2004) The basic reproductive number of Ebola and the effects of public health measures: the cases of Congo and Uganda. J Theor Biol 229(1):119–126
Chowell G, Nishiura H, Bettencourt LM (2007) Comparative estimation of the reproduction number for pandemic influenza from daily case notification data. J R Soc Interface 4(12):155–166
Clarkson JA, Fine PEM (1985) The efficiency of measles and pertussis notification in England and Wales. Int J Epidemiol 14:153–168
Demiris N, O’Neill PD (2006) Computation of final outcome probabilities for the generalised stochastic epidemic. Stat Comput 16(3):309–317
Dorigatti I, Cauchemez S, Pugliese A, Ferguson NM (2012) A new approach to characterising infectious disease transmission dynamics from sentinel surveillance: Application to the Italian 2009–2010 A/H1N1 influenza pandemic. Epidemics 4(1):9–21
Dukic VM, Lopes HF, Polson N (2009) Tracking flu epidemics using google flu trends and particle learning. SSRN. http://ssrn.com/abstract=1513705 or doi:10.2139/ssrn.1513705
Fraser C, Donnelly CA, Cauchemez S, Hanage WP, Van Kerkhove MD, Hollingsworth TD, Griffin J, Baggaley RF, Jenkins HE, Lyons EJ, Jombart T, Hinsley WR, Grassly NC, Balloux F, Ghani AC, Ferguson NM, Rambaut A, Pybus OG, Lopez-Gatell H, Alpuche-Aranda CM, Chapela IB, Zavala EP, Espejo Guevara D Ma, Checchi F, Garcia E, Hugonnet S, Roth C (2009) Pandemic potential of a strain of influenza A (H1N1): early findings. Science 324:1557–1561
Gamado KM, Streftaris G, Zachary S (2014) Modelling under-reporting in epidemics. J Math Biol 69(3):737–765
Gibson GJ, Renshaw E (1998) Estimating parameters in stochastic compartmental models using Markov chain methods. IMA J Math Appl Med Biol 15:19–40
Hastings WK (1970) Monte Carlo sampling using markov chains and their applications. Biometrika 57:97–109
Heesterbeek JAP, Dietz K (1996) The concept of \({R}_0\) in epidemic theory. Stat Neerlandica 50(1):89–110
Hens N, Van Ranst M, Aerts M, Robesyn E, Van Damme P, Beutels P (2011) Estimating the effective reproduction number for pandemic influenza from notification data made publicly available in real time: a multi-country analysis for influenza A/H1N1v 2009. Vaccine 29:896–904
Jewell CP, Keeling MJ, Roberts GO (2008) Predicting undetected infections during the 2007 foot and mouth disease outbreak. JRS Interface 6:1145–1151
Johannes M, Polson NG, Yae SM (2010) Particle learning in nonlinear models using slice variables. Biometrika 1–17. http://faculty.chicagobooth.edu/nicholas.polson/research/papers/Nonli.pdf
Kass RE, Carlin BP, Gelman A, Neal R (1998) Markov chain monte carlo in practice: a roundtable discussion. Am Stat 52:93–100
Knock ES, O’Neill PD (2014) Bayesian model choice for epidemic models with two levels of mixing. Biostatistics (Oxford, England) 15(1):46–59
Legrand J, Grais RF, Boelle PY, Valleron AJ, Flahault A (2007) Understanding the dynamics of Ebola epidemics. Epidemiol Infect 135(4):610–621
Lopes HF, Carvalho CM, Johannes MS, Polson NG (2011) Particle learning for sequential bayesian computation. Bayes Stat 9:317–360
Merler S, Ajelli M, Fumanelli L, Gomes MFC, Piontti APY, Rossi L, Chao DL, Longini IM Jr, Halloran ME, Vespignani A (2015) Spatiotemporal spread of the 2014 outbreak of Ebola virus disease in Liberia and the effectiveness of non-pharmaceutical interventions: a computational modelling analysis. Lancet Infect Dis 15(2):204–211
Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E (1953) Equations of state calculations by fast computing machines. J Chem Phys 21:1087–1091
Neal P, Huang CL (2015) Forward simulation MCMC with applications to stochastic epidemic models. Scand J Stat 42(2):378–396
Neal P, Roberts G (2005) A case study in non-centering for data augmentation: stochastic epidemics. Stat Comput 15:315–327
O’Neill PD, Becker NG (2001) Inference for an epidemic when susceptibility varies. Biostatistics 2(1):99–108
Ortega JM, Rheinboldt WC (2000) Iterative solution of nonlinear equations in several variables (classics in applied mathematics, 30). SIAM, Philadelphia
Papaspiliopoulos O, Roberts GO, Sköld M (2003) Non-centered parameterizations for hierarchical models and data augmentation. In: Bernardo JM, Bayarri MS, Berger JO, Dawid AP, Heckerman D, Smith AFM, West M (eds) Bayesian Statistics 7. Oxford University Press, New York, pp 307–326
Plummer M, Best N, Cowles K, Vines K (2006) Coda: convergence diagnosis and output analysis for mcmc. R News 6(1):7–11
Streftaris G, Gibson G (2004a) Bayesian inference for stochastic epidemics in closed populations. Stat Model 4:63–75
Streftaris G, Gibson GJ (2004b) Bayesian analysis of experimental epidemics of foot-and-mouth disease. Proc R Soc Lond B 271:1111–1117
Streftaris G, Gibson GJ (2012) Non-exponential tolerance to infection in epidemic systems—modelling, inference and assessment. Biostatistics 13(4):580–593
White LF, Pagano M (2010) Reporting errors in infectious disease outbreaks, with an application to pandemic influenza A/H1N1. Epidemiol Perspect Innov 7(1):12
WHO: World Health Organization (2015) Guinea: the ebola virus shows its tenacity. http://www.who.int/csr/disease/ebola/one-year-report/guinea/en/
Xu X, Kypraios T, O’Neill PD (2016) Bayesian non-parametric inference for stochastic epidemic models using gaussian processes. Biostat 1–15. doi:10.1093/biostatistics/kxw011
Acknowledgments
Funding was provided by Heriot-Watt University.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
1.1 A.1 Heuristic justification of the adjustment on p
Considering Eq. (20), we may refer to the following approximation. For a relatively non-informative prior for p, and so also for \({\hat{p}}\), the posterior density of \({\hat{p}}\) is that given by the likelihood function \(L(\cdot )\). Regarding the posterior distributions of \({\hat{p}}\) and p as being modelled by the random variables \({\hat{P}}\) and P respectively, we can define the variable X as
Treating \(\sigma ^2\) as a constant, we have \( X \sim \mathcal {N}(0, \sigma ^2).\) Therefore the multiplicative term \( \phi ({\hat{p}} - p)L(\beta ,\gamma ,{\hat{p}};\,\pmb {s_r}, \pmb {r_r})\), in (20), is the product of the density of X and \({\hat{P}}\). Hence, integrating over \({\hat{p}}\) this gives
which by definition is the convolution of X and \({\hat{P}}\) and, since X and \({\hat{P}}\) are independent, the density of \(P = {\hat{P}} + X\). This result suggests that the means of \({\hat{P}}\) and P are the same and the variance of \({\hat{P}}\) should be increased by the variance of X to obtain the variance of P. In practice the quantity \(\sigma ^2 = p^2(1-p)/K_r\) can be estimated by using any reasonable estimate of p. If we use \({\hat{p}}\) to estimate \(\sigma ^2\), the uncertainty about p can be estimated by adding to the posterior variance of \({\hat{p}}\) the quantity \({\hat{p}}^2(1-\hat{p})/K_r\).
1.2 A.2 Pseudo-code for Methods 1–3
1.2.1 A.2.1 Method 1
MCMC algorithm using Method 1
-
1.
Start with initial values for \(\beta ,\gamma ,p\) and \(\pmb {s_r}\).
-
2.
Update the infection times of the reported cases by selecting one individual at random, propose an infection time based on the distribution of the infectious period and accept it with probability A in (19), given the parameters \(\beta ,\gamma \) and p.
-
3.
Update the model parameters:
-
(i)
Update \(\beta \) according to its full posterior conditional distribution in (17), using the current values of \(p,\gamma \) and \(\pmb {s_r}\).
-
(ii)
Update \(\gamma \) using its full posterior conditional distribution in (18) and the current values of \(p,\beta \) and \(\pmb {s_r}\).
-
(iii)
Update p using random walk with acceptance probability
$$\begin{aligned} A_p = \min \left( \frac{L(\beta ,\gamma , p^{new};\,\pmb {s_r}, \pmb {r_r})}{L(\beta ,\gamma , p^{old};\,\pmb {s_r}, \pmb {r_r})}, 1\right) \end{aligned}$$using current \(\gamma ,\beta \) and \(\pmb {s_r}\).
-
(i)
-
(iv)
Steps 2 and 3 are repeated until convergence.
1.2.2 A.2.2 Method 2
Algorithm for correction on p
-
1.
Start with initial values for \(\beta ,\gamma ,{\hat{p}},p\) and \(\pmb {s_r}\).
-
2.
Update the infection times of the reported cases, \(\pmb {s_r}\), using Metropolis–Hastings algorithm, given the current values of parameters \(\beta ,\gamma ,{\hat{p}}\) and p.
-
3.
Update the model parameters:
-
(i)
Update \(\beta \) according to its full posterior conditional distribution in (17) with p replaced by \({\hat{p}}\), using the current values of \(p,{\hat{p}},\gamma \) and \(\pmb {s_r}\).
-
(ii)
Update \(\gamma \) using its full posterior conditional distribution in (18) and the current values of \(p,{\hat{p}},\beta \) and \(\pmb {s_r}\).
-
(iii)
Update \({\hat{p}}\) following a random walk scheme with acceptance probability
$$\begin{aligned} Acc_{{\hat{p}}} = \min \left( \frac{\phi ({\hat{p}}^{new} - p)L(\beta ,\gamma ,{\hat{p}}^{new};\,\pmb {s_r}, \pmb {r_r})}{\phi (\hat{p}^{old} - p) L(\beta ,\gamma ,{\hat{p}}^{old};\,\pmb {s_r}, \pmb {r_r})}, 1\right) \end{aligned}$$using the current values of \(p,\gamma ,\beta \) and \(\pmb {s_r}\).
-
(iv)
Update p using random walk with acceptance probability
$$\begin{aligned} Acc_p = \min \left( \frac{\pi _p(p^{new}) \phi ({\hat{p}} - p^{new})}{\pi _p(p^{old})\phi ({\hat{p}} - p^{old})}, 1\right) \end{aligned}$$using \({\hat{p}},\gamma ,\beta \) and \(\pmb {s_r}\). Note that this update is actually independent of \(\beta ,\gamma \) and \(\pmb {s_r}\).
-
(i)
-
4.
Steps 2 and 3 are repeated until convergence.
1.2.3 A.2.3 Method 3
Algorithm for approximate Gibbs sampling
-
1.
Start with initial values of \(\beta ,\gamma ,p\) and \(\pmb {s_r}\).
-
2.
Update the infection times of the reported cases, \(\pmb {s_r}\), using Metropolis–Hastings algorithm, given the current values of parameters \(\beta ,\gamma \) and p.
-
3.
Update the model parameters:
-
(i)
Update \(\beta \) according to its full posterior conditional distribution in (17) using the current values of \(p,\gamma \) and \(\pmb {s_r}\).
-
(ii)
Update \(\gamma \) using its full posterior conditional distribution in (18) and the current values of \(p,\beta \) and \(\pmb {s_r}\).
-
(iii)
Solve Eq. (13) and sample the final size K from (14) using current \(\beta ,\gamma \) and \(\pmb {s_r}\); conditional on the fact that \(K \ge K_r\).
-
(iv)
Sample the probability of reporting p using (23) using the current final size K.
-
(i)
-
4.
Steps 2 and 3 are repeated until convergence.
1.3 A.3 Reversible jump MCMC algorithm
An individual (say k) will always have one of the following states:
-
0: Susceptible;
i.e \(k \in \mathcal {S}\)
-
1: Removed and reported;
i.e \(k \in \mathcal {R}_r\)
-
2: Removed but not reported;
i.e \(k \in \mathcal {R}_u\).
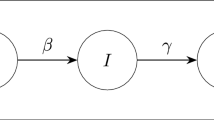
Below are the possible algorithm transitions presented schematically:

The algorithm is now describe in details as follows:
-
Choose an individual at random (let us say k).
-
If the state of k is 1 (meaning that the individual was removed and reported), we propose the new infection time \(s_k\) using \((s_k - r_k) \sim \text {Exp}(\gamma )\) and \(s_k\) is accepted with probability :
$$\begin{aligned} A_{1\rightarrow 1} = \min \left\{ 1, \frac{L(\beta , \gamma ; \pmb {s}^{(new)}, \pmb {r})}{L(\beta , \gamma ; \pmb {s}^{(old)}, \pmb {r})}*\frac{\exp \{-\gamma (r_k - s_k')\}}{\exp \{-\gamma (r_k - s_k)\}}\right\} \end{aligned}$$where \(s_k'\) is the current infection time of the individual k. There is no change in state.
-
If the state of k is 0 (susceptible individual) we propose a removal time \(r_k\) uniformly in \((T_0, T)\) (the interval \((T_0, T)\) is the time window in which the epidemic has occurred) and an infection time \(s_k\) in \((T_0, r_k)\) and add the pair with probability
$$\begin{aligned} A_{0\rightarrow 2} = \min \left\{ 1, \frac{(T-T_0)(r_k - T_0)}{2}\frac{L(\beta , \gamma , p; \pmb {s}^{(new)}, \pmb {r}^{(new)})}{L(\beta , \gamma , p; \pmb {s}^{(old)}, \pmb {r}^{(old)})}\right\} . \end{aligned}$$If this move is accepted, the state of the individual k becomes 2 (which represents individuals that are removed but not reported), i.e
$$\begin{aligned} |S| = |S| - 1 \quad \text { and } \quad |\mathcal {R}_u| = |\mathcal {R}_u| + 1. \end{aligned}$$ -
If state of k is 2 we either propose, with probability 1 / 2, to update the pair of infection and removal times, or delete the pair of infection and removal times:
-
Update the pair of infection and removal times of k (with \(T_0 \le s_k < r_k \le T\)) with probability
$$\begin{aligned} A_{2\rightarrow 2} = \min \left\{ 1, \frac{L(\beta , \gamma , p; \pmb {s}^{(new)}, \pmb {r})}{L(\beta , \gamma , p; \pmb {s}^{(old)}, \pmb {r})} \frac{r_k - T_0}{r_k' - T_0}\right\} \end{aligned}$$where \(r_k'\) is the removal time of individual k before the new proposed one \(r_k\). The state remains the same.
-
Delete the pair of infection and removal times with probability
$$\begin{aligned} A_{2\rightarrow 0} = \min \left\{ 1, \frac{L(\beta , \gamma , p; \pmb {s}^{(new)}, \pmb {r})}{L(\beta , \gamma , p; \pmb {s}^{(old)}, \pmb {r})} \frac{2}{(T - T_0)(r_k - T_0)}\right\} . \end{aligned}$$The state of the individual k becomes 0 if the deletion is accepted, i.e
$$\begin{aligned} |\mathcal {R}_u| = |\mathcal {R}_u| - 1 \qquad \text { and } \qquad |\mathcal {S}| = |\mathcal {S}| + 1. \end{aligned}$$
-
1.4 A.4 Auto-correlation functions
Auto-correlation function for parameter \(\gamma \) (data from Sect. 4). a ACF plot for \(\gamma \), Method 1. b ACF plot for \(\gamma \), Method 2. c ACF plot for \(\gamma \), Method 3. d ACF plot for \(\gamma \), Method 4
Auto-correlation function for parameter K (data from Sect. 4). a ACF plot for K, Method 1. b ACF plot for K, Method 2. c ACF plot for K, Method 3. d ACF plot for K, Method 4
Rights and permissions
About this article
Cite this article
Gamado, K., Streftaris, G. & Zachary, S. Estimation of under-reporting in epidemics using approximations. J. Math. Biol. 74, 1683–1707 (2017). https://doi.org/10.1007/s00285-016-1064-7
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-016-1064-7
Keywords
- Under-reporting
- Approximations
- Markov chain Monte Carlo
- Reversible jump
- Stochastic SIR model
- Final size distribution