Abstract
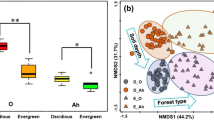
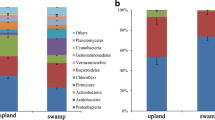
Soil microbial communities are engineers of important biogeochemical processes and play a critical role in regulating the functions and stability of forest ecosystem. However, few studies have assessed microbial interactions during forest conversion, which is essential to the understanding of the structure and function of soil microbiome. Herein, we investigated the co-occurrence network pattern and putative functions of fungal and bacterial communities in forest-transforming areas (five sites that cover the typical forests) using high-throughput sequencing of the ITS genes and 16S rRNA. Our study showed that the bacterial network had higher average connectivity and more links than fungal network, which might indicate that the bacterial community had more complex internal interactions compared with fungal one. Alphaproteobacteria_unclassfied, Telmatobacter, 0319-6A21 and Latescibacteria_unclassfied were the keystone taxa in bacterial network. For the fungal community network, the keystone taxon was Ceratobasidium. A structural equation model indicated that the available potassium and total organic carbon were important soil environmental factors, which affected all microbial modules, including bacterial and fungi. Total nitrogen had significant effects on the bacterial module that contains a relatively rich group of nitrogen cycling functions, and pH influenced the bacterial module which have higher potential functions of carbon cycling. And, more fungal modules were directly affected by forest structure (S Tree) compared with bacterial ones. This study provides new insights into our understanding of the feedback of underground creatures to forest conversion and highlights the importance of microbial modules in the nutrient cycling process.





Similar content being viewed by others
Data Availability
ITS genes and 16S rRNA Illumina sequence reads were deposited in NCBI SRA under BioProject ID PRJNA388530.
References
Guo LB, Gifford RM (2002) Soil carbon stocks and land use change: a meta analysis. Global Chang Biol 8:345–360. https://doi.org/10.1046/j.1354-1013.2002.00486.x
Hardwick SR, Toumi R, Pfeifer M, Turner EC, Nilus R, Ewers RM (2015) The relationship between leaf area index and microclimate in tropical forest and oil palm plantation: Forest disturbance drives changes in microclimate. Agric For Meteorol 201:187–195. https://doi.org/10.1016/j.agrformet.2014.11.010
Ozalp M, Erdogan Yuksel E, Yuksek T (2016) Soil property changes after conversion from forest to pasture in Mount Sacinka, Artvin, Turkey. Land Degrad Dev 27:1007–1017. https://doi.org/10.1002/ldr.2353
Bowd EJ, Banks SC, Strong CL, Lindenmayer DB (2019) Long-term impacts of wildfire and logging on forest soils. Nat Geosci 12:113–118. https://doi.org/10.1038/s41561-018-0294-2
Jin X, Liu Y, Hu W, Wang G, Kong Z, Wu L, Ge G (2019) Soil bacterial and fungal communities and the associated nutrient cycling responses to forest conversion after selective logging in a subtropical forest of China. For Ecol Manage 444:308–317. https://doi.org/10.1016/j.foreco.2019.04.032
Bender SF, Wagg C, van der Heijden MG (2016) An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol Evol 31:440–452. https://doi.org/10.1016/j.tree.2016.02.016
Hartmann M, Niklaus PA, Zimmermann S, Schmutz S, Kremer J, Abarenkov K, Lüscher P, Widmer F, Frey B (2014) Resistance and resilience of the forest soil microbiome to logging-associated compaction. ISME J 8:226–244. https://doi.org/10.1038/ismej.2013.141
Wood SA, Gilbert JA, Leff JW, Fierer N, D’Angelo H, Bateman C, Gedallovich SM, Gillikin CM, Gradoville MR, Mansor P (2017) Consequences of tropical forest conversion to oil palm on soil bacterial community and network structure. Soil Biol Biochem 112:258–268. https://doi.org/10.1016/j.soilbio.2017.05.019
Tripathi BM, Edwards DP, Mendes LW, Mincheol K, Ke D, Hyoki K, Adams JM (2016) The impact of tropical forest logging and oil palm agriculture on the soil microbiome. Mol Ecol 25(10):2244–2257. https://doi.org/10.1111/mec.13620
Faust K, Raes J (2012) Microbial interactions: from networks to models. Nat Rev Microbiol 10(8):538. https://doi.org/10.1038/nrmicro2832
Delgado-Baquerizo M, Reith F, Dennis PG, Hamonts K, Powell JR, Young A, Singh BK, Bissett A (2018) Ecological drivers of soil microbial diversity and soil biological networks in the Southern Hemisphere. Ecology 99(3):583–596. https://doi.org/10.1002/ecy.2137
Shi Y, Li Y, Xiang X, Sun R, Yang T, He D, Zhang K, Ni Y, Zhu Y-G, Adams JM (2018) Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain. Microbiome 6:27. https://doi.org/10.1186/s40168-018-0409-4
Menezes ABD, Prendergast-Miller MT, Richardson AE, Toscas P, Farrell M, Macdonald LM, Baker G, Wark T, Thrall PH (2015) Network analysis reveals that bacteria and fungi form modules that correlate independently with soil parameters. Environ Microbiol 17:2677–2689. https://doi.org/10.1111/1462-2920.12559
Banerjee S, Schlaeppi K, van der Heijden MG (2018) Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol 16:567–576. https://doi.org/10.1038/s41579-018-0024-1
Berry D, Widder S (2014) Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol 5:219. https://doi.org/10.3389/fmicb.2014.00219
Herren CM, McMahon KD (2018) Keystone taxa predict compositional change in microbial communities. Environ Microbiol 20:2207–2217. https://doi.org/10.1111/1462-2920.14257
Zhou H, Gao Y, Jia X, Wang M, Ding J, Cheng L, Bao F, Wu B (2020) Network analysis reveals the strengthening of microbial interaction in biological soil crust development in the Mu Us Sandy Land, northwestern China. Soil Biol Biochem 144:107782. https://doi.org/10.1016/j.soilbio.2020.107782
Thiergart T, Duran P, Ellis T, Garrido-Oter R, Kemen E, Roux F, Alonso-Blanco C, Ågren J, Schulze-Lefert P, Hacquard S (2020) Root microbiota assembly and adaptive differentiation among European Arabidopsis populations. Nat Ecol Evol 4:122–131. https://doi.org/10.1038/s41559-019-1063-3
Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO (2013) The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:643–648. https://doi.org/10.1093/nar/gkt1209
Wang K, Yin X, Mao H, Chu C, Tian Y (2018) Changes in structure and function of fungal community in cow manure composting. Bioresour Technol 255:123–130. https://doi.org/10.1016/j.biortech.2018.01.064
Langfelder P, Horvath S (2012) Fast R functions for robust correlations and hierarchical clustering. J Stat Softw 46:11. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3465711/?report=classic
Luo F, Yang Y, Zhong J, Gao H, Khan L, Thompson DK, Zhou J (2007) Constructing gene co-expression networks and predicting functions of unknown genes by random matrix theory. BMC Bioinformatics 8:299. https://doi.org/10.1186/1471-2105-8-299
Hu HW, Wang JT, Li J, Li JJ, Ma YB, Chen D, He JZ (2016) Field-based evidence for copper contamination induced changes of antibiotic resistance in agricultural soils. Environ Microbiol 18:3896–3909. https://doi.org/10.1111/1462-2920.13370
Bastian M, Heymann S, Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks. Third international AAAI conference on weblogs and social media. https://doi.org/10.13140/2.1.1341.1520
Deng Y, Jiang YH, Yang Y, He Z, Luo F, Zhou J (2012) Molecular ecological network analyses. BMC Bioinformatics 13:113. https://doi.org/10.1186/1471-2105-13-113
Guimera R, Amaral LAN (2005) Functional cartography of complex metabolic networks. Nature 433:895–900. https://doi.org/10.1038/nature03288
Olesen JM, Bascompte J, Dupont YL, Jordano P (2007) The modularity of pollination networks. Proc Natl Acad Sci USA 104:19891–19896. https://doi.org/10.1073/pnas.0706375104
Louca S, Parfrey LW, Doebeli M (2016) Decoupling function and taxonomy in the global ocean microbiome. Science 353:1272–1277. https://doi.org/10.1126/science.aaf4507
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248. https://doi.org/10.1016/j.funeco.2015.06.006
Qi L, Yang J (2017) Microbial community composition regulates SOC decomposition response to forest conversion in a Chinese temperate forest. Ecol Res 32:1–10. https://doi.org/10.1007/s11284-016-1428-x
Pei Z, Leppert KN, Eichenberg D, Bruelheide H, Niklaus PA, Buscot F, Gutknecht JLM (2017) Leaf litter diversity alters microbial activity, microbial abundances, and nutrient cycling in a subtropical forest ecosystem. Biogeochemistry 134:163–181. https://doi.org/10.1007/s10533-017-0353-6
Shipley B (2013) The AIC model selection method applied to path analytic models compared using a d-separation test. Ecology 94:560–564. https://doi.org/10.1890/12-0976.1
Coyte KZ, Schluter J, Foster KR (2015) The ecology of the microbiome: networks, competition, and stability. Science 350:663–666. https://doi.org/10.1126/science.aad2602
Banerjee S, Kirkby CA, Schmutter D, Bissett A, Kirkegaard JA, Richardson AE (2016) Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol Biochem 97:188–198. https://doi.org/10.1016/j.soilbio.2016.03.017
Bissett A, Brown MV, Siciliano SD, Thrall PH (2013) Microbial community responses to anthropogenically induced environmental change: towards a systems approach. Ecol Lett 16:128–139. https://doi.org/10.1111/ele.12109
Pan YQ, Kang P, Hu JP, Song NP (2021) Bacterial community demonstrates stronger network connectivity than fungal community in desert-grassland salt marsh. Sci Total Environ 798:149118. https://doi.org/10.1016/j.scitotenv.2021.149118
Eiler A, Heinrich F, Bertilsson S (2012) Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J 6:330–342. https://doi.org/10.1038/ismej.2011.113
Wu L, Yang Y, Chen S, Zhao M, Zhu Z, Yang S, Qu Y, Ma Q, He Z, Zhou J (2016) Long-term successional dynamics of microbial association networks in anaerobic digestion processes. Water Res 104:1–10. https://doi.org/10.1016/j.watres.2016.07.072
Tian J, He N, Kong W, Deng Y, Feng K, Green SM, Wang X, Zhou J, Kuzyakov Y, Yu G (2018) Deforestation decreases spatial turnover and alters the network interactions in soil bacterial communities. Soil Biol Biochem 123:80–86. https://doi.org/10.1016/j.soilbio.2018.05.007
Thomson BC, Ostle N, McNamara N, Bailey MJ, Whiteley AS, Griffiths RI (2010) Vegetation affects the relative abundances of dominant soil bacterial taxa and soil respiration rates in an upland grassland soil. Microb Ecol 59(2):335–343. https://doi.org/10.1007/s00248-009-9575-z
Zhou ZH, Wang CK (2018) Effects of forest degradation on microbial communities and soil carbon cycling: A global meta-analysis. Global Ecol Biogeogr 27:110–124. https://doi.org/10.1111/geb.12663
Singh BK, Bardgett RD, Smith P, Reay DS (2010) Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol 8(11):779. https://doi.org/10.1038/nrmicro2439
Zhang X, Liu S, Huang Y, Fu S, Wang J, Ming A, Li X, Yao M, Li H (2018) Tree species mixture inhibits soil organic carbon mineralization accompanied by decreased r-selected bacteria. Plant Soil 431:203–216. https://doi.org/10.1007/s11104-018-3755-x
Khan IU, Hussain F, Habib N, Wadaan MA, Ahmed I, Im W-T, Hozzein WN, Zhi X-Y, Li W-J (2017) Phenylobacterium deserti sp nov, isolated from desert soil. Int J Syst Evol Microbiol 67(11):4722–4727. https://doi.org/10.1099/ijsem.0.002366
Lladó S, Žifčáková L, Větrovský T, Eichlerová I, Baldrian P (2016) Functional screening of abundant bacteria from acidic forest soil indicates the metabolic potential of Acidobacteria subdivision 1 for polysaccharide decomposition. Biol Fertil Soils 52(2):251–260. https://doi.org/10.1007/s00374-015-1072-6
Chang Y-J, Land M, Hauser L, Chertkov O, Del Rio TG, Nolan M, Copeland A, Tice H, Cheng J-F, Lucas S (2011) Non-contiguous finished genome sequence and contextual data of the filamentous soil bacterium Ktedonobacter racemifer type strain (SOSP1-21 T). Stand Genomic Sci 5:97. https://doi.org/10.4056/sigs.2114901
Lange L, Pilgaard B, Herbst F-A, Busk PK, Gleason F, Pedersen AG (2019) Origin of fungal biomass degrading enzymes: evolution. diversity and function of enzymes of early lineage fungi. Fungal Biol Rev 33:82–97. https://doi.org/10.1016/j.fbr.2018.09.001
Fernandez CW, Kennedy PG (2016) Revisiting the ‘Gadgil effect’: do interguild fungal interactions control carbon cycling in forest soils? New Phytol 209(4):1382–1394. https://doi.org/10.1111/nph.13648
Montoya JM, Pimm SL, Solé RV (2006) Ecological networks and their fragility. Nature 442(7100):259. https://doi.org/10.1038/nature04927
Tao J, Meng D, Qin C, Liu X, Liang Y, Xiao Y, Liu Z, Gu Y, Li J, Yin H (2018) Integrated network analysis reveals the importance of microbial interactions for maize growth. Appl Microbiol Biot 102(8):3805–3818. https://doi.org/10.1007/s00253-018-8837-4
Youssef NH, Farag IF, Rinke C, Hallam SJ, Woyke T, Elshahed MS (2015) In Silico analysis of the metabolic potential and niche specialization of candidate phylum" Latescibacteria"(WS3). PLoS ONE 10:e0127499. https://doi.org/10.1371/journal.pone.0127499
Pankratov TA, Kirsanova LA, Kaparullina EN, Kevbrin VV, Dedysh SN (2012) Telmatobacter bradus gen nov, sp nov, a cellulolytic facultative anaerobe from subdivision 1 of the Acidobacteria, and emended description of Acidobacterium capsulatum Kishimoto et al. 1991. Int J Syst Evol Microbiol 62:430–437. https://doi.org/10.1099/ijs.0.029629-0
Swift S, Munroe S, Im C, Tipton L, Hynson NA (2019) Remote tropical island colonization does not preclude symbiotic specialists: new evidence of mycorrhizal specificity across the geographic distribution of the Hawaiian endemic orchid Anoectochilus sandvicensis. Ann Bot 123:657–666. https://doi.org/10.1093/aob/mcy198
Zhong ZK, Zhang XY, Wang X, Fu SY, Wu SJ, Lu XQ, Ren CJ, Han XH, Yang GH (2020) Soil bacteria and fungi respond differently to plant diversity and plant family composition during the secondary succession of abandoned farmland on the Loess Plateau. China Plant Soil 448(1):183–200. https://doi.org/10.1007/s11104-019-04415-0
Dassen S, Cortois R, Martens H, de Hollander M, Kowalchuk GA, van der Putten WH, De Deyn GB (2017) Differential responses of soil bacteria, fungi, archaea and protists to plant species richness and plant functional group identity. Mol Ecol 26:4085–4098. https://doi.org/10.1111/mec.14175
Vukicevich E, Lowery T, Bowen P, Úrbez-Torres JR, Hart M (2016) Cover crops to increase soil microbial diversity and mitigate decline in perennial agriculture. A review Agron Sustain Dev 36:48. https://doi.org/10.1007/s13593-016-0385-7
Zuo Y, He C, He X, Li X, Xue Z, Li X, Wang S (2020) Plant cover of Ammopiptanthus mongolicus and soil factors shape soil microbial community and catabolic functional diversity in the arid desert in Northwest China. Appl Soil Ecol 147:103389. https://doi.org/10.1016/j.apsoil.2019.103389
Malik AA, Puissant J, Buckeridge KM, Goodall T, Jehmlich N, Chowdhury S, Gweon HS, Peyton JM, Mason KE, van Agtmaal M, Blaud A, Clark IM, Whitaker J, Pywell RF, Ostle N, Gleixner G, Griffiths RI (2018) Land use driven change in soil pH affects microbial carbon cycling processes. Nat Commun 9:3591–3591. https://doi.org/10.1038/s41467-018-05980-1
Cassman NA, Leite MFA, Pan Y, de Hollander M, van Veen JA, Kuramae EE (2016) Plant and soil fungal but not soil bacterial communities are linked in long-term fertilized grassland. Scientific Rep-UK 6:23680. https://doi.org/10.1038/srep23680
Clayton J, Lemanski K, Bonkowski M (2021) Shifts in soil microbial stoichiometry and metabolic quotient provide evidence for a critical tipping point at 1% soil organic carbon in an agricultural post-mining chronosequence. Biol Fert Soils 57:435–446. https://doi.org/10.1007/s00374-020-01532-2
Li JW, Shangguan ZP, Deng L (2020) Variations of belowground C and N cycling between arbuscular mycorrhizal and ectomycorrhizal forests across China. Soil Res 58:441–445. https://doi.org/10.1071/SR19377
Bastida F, Eldridge DJ, García C, Kenny Png G, Bardgett RD, Delgado-Baquerizo M (2021) Soil microbial diversity-biomass relationships are driven by soil carbon content across global biomes. ISME J 15:2081–2091. https://doi.org/10.1038/s41396-021-00906-0
Řezáčová V, Konvalinková T, Jansa J (2017) Carbon fluxes in mycorrhizal plants. In Mycorrhiza-eco-physiology, secondary metabolites, nanomaterials Springer Cham. https://doi.org/10.1007/978-3-319-57849-1_1
Acknowledgements
This work was supported by the National Natural Science Foundations of China (Grant No. 31560143, 31660149 and 31971470). The authors thank Ruichang Shen (Nanchang University) for examination of a part of the experimental conditions; Yantian Ma (Nanchang University) for helpful discussions; We also wish to thank Mr. Guobing Wang and Mr. Zeping Yu of the Jiangxi Guanshan National Nature Reserve for his assistance with the sampling in the field.
Funding
This work was supported by the National Natural Science Foundations of China (Grant No. 31560143, 31660149 and 31971470).
Author information
Authors and Affiliations
Contributions
Study concept and design: GG and LW. Analysis and interpretation of data: YL and XJ. Drafting of the manuscript: YL. Critical revision of the manuscript for important intellectual content: ZK and YL. Statistical analysis: SH. Obtained funding: GG. Study supervision: LW.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
The submitted work is original and have not been submitted to other journals.
Consent to Participate
The authors have agreed to be listed and approved the submitted version of the manuscript.
Informed Consent
The authors hereby consent for the article publication in Current Microbiology journal.
Research Involving Human and Animal Participants
This article does not contain any studies with humans or animal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, Y., Jin, X., Huang, S. et al. Co-Occurrence Patterns of Soil Fungal and Bacterial Communities in Subtropical Forest-Transforming Areas. Curr Microbiol 81, 64 (2024). https://doi.org/10.1007/s00284-023-03608-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03608-2