Abstract
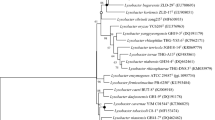
A bacterium, designated 50T was isolated from the sediment of a pesticide plant in Shandong Province, PR China. The strain was non-motile, Gram stain-negative, rod shaped and grew optimally on NA medium at 30 °C, pH 7.5 and with 0% (w/v) NaCl. Strain 50T showed the highest 16S rRNA gene sequence similarity with Lysobacter pocheonensis Gsoil 193T (96.7%), followed by Luteimonas lumbrici 1.1416T (96.5%). Phylogenetic analyses based on 16S rRNA indicated that strain 50T and Luteimonas lumbrici 1.1416T were clustered with the genus of Lysobacter and formed a subclade with Lysobacter pocheonensis Gsoil 193T. In the phylogenetic analysis based on the genome sequences, strain 50T and Luteimonas lumbrici 1.1416T were also clustered with the type strains of the genus Lysobacter. The obtained ANI and the dDDH value between 50T and Luteimonas lumbrici 1.1416T were 80.6% and 24.0%, respectively. The respiratory quinone was ubiquinone-8 (Q-8), and the major cellular fatty acids were iso-C15: 0 (31.7%), summed feature 9 (iso-C17:1 ω9c or C16:0 10-methyl) (23.7%), iso-C17:0 (14.3%) and iso-C16:0 (12.6%). The major polar lipids were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine and unidentified aminophospholipid, unidentified phospholipid and unidentified lipid. The genomic DNA G + C content was 69.5 mol%. According to the phenotypic, chemotaxonomic and phylogenetic analyses, strain 50T represents a novel species of the genus Lysobacter, for which the name Lysobacter sedimenti sp. nov. is proposed, with strain 50T (= KCTC 92088T = CCTCC AB 2022035T) as the type strain. In this study, it is also proposed that Luteimonas lumbrici should be transferred to the genus Lysobacter as Lysobacter lumbrici comb. nov. The type strain of Lysobacter lumbrici is 1.1416T (= KCTC 62979T = CCTCC AB 2018348T).

Similar content being viewed by others
Data Availability
All authors have declared that all data are available.
References
Penelope C, Cook D (1978) Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. Int J Syst Evol Microbiol 28(3):367–393. https://doi.org/10.1099/00207713-28-3-367
Yoon JH, Lee ST, Park YH (1998) Inter- and intraspecific phylogenetic analysis of the genus Nocardioides and related taxa based on 16S rDNA sequences. Int J Syst Bacteriol 48(Pt 1):187–194. https://doi.org/10.1099/00207713-48-1-187
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M et al (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62(Pt 3):716–721. https://doi.org/10.1099/ijs.0.038075-0
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17(6):368–376. https://doi.org/10.1007/bf01734359
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20(4):406–416. https://doi.org/10.1093/sysbio/20.4.406
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120. https://doi.org/10.1007/bf01731581
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Chaudhari NM, Gupta VK, Dutta C (2016) BPGA- an ultra-fast pan-genome analysis pipeline. Sci Rep 6:24373. https://doi.org/10.1038/srep24373
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17(4):540–552. https://doi.org/10.1093/oxfordjournals.molbev.a026334
Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32(1):268–274. https://doi.org/10.1093/molbev/msu300
Green MR, Sambrook J (2001) Molecular cloning: a laboratory manual. Anal Biochem 186(1):182–183
Luo R, Liu B, Xie Y, Li Z, Huang W et al (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1(1):18. https://doi.org/10.1186/2047-217x-1-18
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. https://doi.org/10.1186/1471-2164-9-75
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ et al (2014) The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:206–214. https://doi.org/10.1093/nar/gkt1226
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68(1):461–466. https://doi.org/10.1099/ijsem.0.002516
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. MIDI Inc, Newark
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of cellulomonas, oerskovia and related taxa. J Appl Bacteriol 47(1):87–95. https://doi.org/10.1111/j.1365-2672.1979.tb01172.x
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M et al (1984) An integrated procedure for the extration of bacterial isoprenoid quinones and polar lipids. Elsevier 2:233–241. https://doi.org/10.1016/0167-7012(84)90018-6
Komagata K (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106(45):19126–19131. https://doi.org/10.1073/pnas.0906412106
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60. https://doi.org/10.1186/1471-2105-14-60
Cha QY, Zhou XK, Zhang XF, Li M, Wei YQ et al (2020) Luteimonas lumbrici sp. nov., a novel bacterium isolated from wormcast. Int J Syst Evol Microbiol 70(1):604–610. https://doi.org/10.1099/ijsem.0.003799
Siddiqi MZ, Im WT (2016) Lysobacter pocheonensis sp. nov., isolated from soil of a ginseng field. Arch Microbiol 198(6):551–557. https://doi.org/10.1007/s00203-016-1214-8
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31800097 and 32170128).
Author information
Authors and Affiliations
Contributions
Conceptualization, funding acquisition and supervision: CC and JH; Laboratory work, data analysis and writing of the original draft: XZ; Writing, reviewing and editing of the manuscript: NW, KG and YP. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. All the authors declare that they have no conflict of interest.
Ethical Approval
The authors have declared that no ethical issues exist.
Research Involving Human and/or Animal Participants
This article does not contain any studies with human participants or animals performed by any of the author.
Consent to Participate and Consent for Publication
All authors agree to have participated in the research proposed to be published and agree to be published in the journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences and the whole genome of strain 50T are ON037515 and JALGCL000000000, respectively.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Wu, N., Geng, K. et al. Lysobacter sedimenti sp. nov., Isolated from the Sediment, and Reclassification of Luteimonas lumbrici as Lysobacter lumbrici comb. nov. Curr Microbiol 79, 381 (2022). https://doi.org/10.1007/s00284-022-03084-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03084-0