Abstract
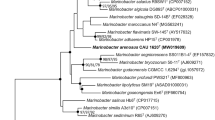
A Gram-negative, facultatively anaerobic, motile, and rod-shaped bacterium, designated ASW11-47 T, was isolated from a tidal flat sediment taken from the coast of Qingdao, PR China. Phylogenetic analysis of 16S rRNA gene sequence showed that strain ASW11-47 T belongs to the genus Salinimicrobium and is most closely related to Salinimicrobium terrae YIM-C338T (98.68% similarity). The length of draft genome is 3,594,457 bp, and DNA G + C content is 40.8 mol%. The values of average nucleotide identity and digital DNA–DNA hybridization between strain ASW11-47 T and closely related strains were in ranges of 75.9–85.9 and 19.7–31.5%, respectively. The major fatty acids (> 10%) were iso-C15:0 and iso-C17:0 3-OH. The predominant respiratory quinone was menaquinone-6 and the major polar lipid was phosphatidylethanolamine. On the basis of genotypic, phenotypic, and chemotaxonomic analysis, strain ASW11-47 T represents a novel species within the genus Salinimicrobium, for which the name Salinimicrobium sediminilitoris sp. nov. is proposed. The type strain is ASW11-47 T (= KCTC 82501 T = MCCC 1K05586T).

Similar content being viewed by others
Data Availability
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequence and the genome sequence of strain ASW11-47 T are OK161349 and JAIVCC000000000, respectively.
Abbreviations
- ANI:
-
Average nucleotide identity
- dDDH:
-
Digital DNA–DNA hybridization
References
Garrity GM, Holt JG (2001) The road map to the manual. In: Garrity GM, Boone DR, Castenholz RW (eds) Bergey’s Manual of Systematic Bacteriology, 2nd edn. Springer, New York, pp 119–166
Kirchman DL (2002) The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol 39:91–100. https://doi.org/10.1111/j.1574-6941.2002.tb00910.x
Cottrell MT, Kirchman DL (2000) Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol 66:1692–1697. https://doi.org/10.1128/AEM.66.4.1692-1697.2000
Lim JM, Jeon CO, Lee SS, Park DJ, Xu LH, Jiang CL, Kim CJReclassification of Salegentibacter catena Ying, et al (2008) 2007 as Salinimicrobium catena gen. nov., comb. nov. and description of Salinimicrobium xinjiangense sp. nov., a halophilic bacterium isolated from Xinjiang province in China. Int J Syst Evol Microbiol 58:438–442. https://doi.org/10.1099/ijs.0.65297-0
Ying JY, Liu ZP, Wang BJ, Dai X, Yang SS, Liu SJ (2007) Salegentibacter catena sp. nov., isolated from sediment of the South China sea, and emended description of the genus Salegentibacter. Int J Syst Evol Microbiol 57:219–222. https://doi.org/10.1099/ijs.0.64658-0
Nedashkovskaya OI, Vancanneyt M, Kim SB, Han J, Zhukova NV, Shevchenko LS (2010) Salinimicrobium marinum sp. nov., a halophilic bacterium of the family Flavobacteriaceae, and emended descriptions of the genus Salinimicrobium and Salinimicrobium catena. Int J Syst Evol Microbiol 60:2303–2306. https://doi.org/10.1099/ijs.0.019166-0
Lee SY, Park S, Oh TK, Yoon JH (2012) Salinimicrobium gaetbulicola sp. nov., isolated from tidal flat sediment. Int J Syst Evol Microbiol 62:1027–1031. https://doi.org/10.1099/ijs.0.033399-0
Subhash Y, Sasikala C, Ramana CV (2014) Salinimicrobium sediminis sp. nov., isolated from a deep-sea sediment. Int J Syst Evol Microbiol 64:984–988. https://doi.org/10.1099/ijs.0.058149-0
Zhang H, Chang YQ, Zheng WS, Chen GJ, Du ZJ (2017) Salinimicrobium flavum sp. nov., isolated from coastal sediment. Int J Syst Evol Microbiol 67:4083–4088. https://doi.org/10.1099/ijsem.0.002257
Cao WR, Zhang LZ, Hu YH, Jiang MY, Li YJ (2020) Salinimicrobium nanhaiense sp. nov. and Salinimicrobium oceani sp. nov. two novel species of the family Flavobacteriaceae isolated from the South China Sea. Int J Syst Evol Microbiol 70:5263–5270. https://doi.org/10.1099/ijsem.0.004405
Chen YG, Cui XL, Zhang YQ, Li WJ, Wang YX, Kim CJ, Lim JM, Xu LH, Jiang CL (2008) Salinimicrobium terrae sp. nov., isolated from saline soil, and emended description of the genus Salinimicrobium. Int J Syst Evol Microbiol 58:2501–2504. https://doi.org/10.1099/ijs.0.65860-0
Kim JH, Yoon JH, Kim W (2016) Salinimicrobium soli sp. nov., isolated from soil of reclaimed land. Int J Syst Evol Microbiol 66:462–467. https://doi.org/10.1099/ijsem.0.000741
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acids techniques in bacterial systematics. Wiley, Chichester, pp 115–147
Li Y, Ding YY, Dang YR, Bai Y, Guan L, Liu NH, Wang YZ, Kang ML, Zhang YQ, Zhang XY (2022) Celeribacter litoreus sp. nov., isolated from intertidal sediment. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.005241
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416. https://doi.org/10.1093/sysbio/20.4.406
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Jackman SD, Vandervalk BP, Mohamadi H, Chu J, Yeo S, Hammond SA, Jahesh G, Khan H, Coombe L, Warren RL, Birol I (2017) ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res 27:768–777. https://doi.org/10.1101/gr.214346.116
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. https://doi.org/10.1186/1471-2164-9-75
Haft DH, DiCuccio M, Badretdin A, Brover V, Chetvernin V, O’Neill K, Li WJ, Chitsaz F, Derbyshire MK, Gonzales NR, Gwadz M, Lu F, Marchler GH, Song JS, Thanki N, Yamashita RA, Zheng CJ, Thibaud-Nissen F, Geer LY, Marchler-Bauer A, Pruitt KD (2018) RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res 46:D851–D860. https://doi.org/10.1093/nar/gkx1068
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. https://doi.org/10.1093/nar/gkw569
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf 14:60. https://doi.org/10.1186/1471-2105-14-60
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. https://doi.org/10.1093/bioinformatics/8.3.275
Buck JD (1982) Nonstaining (KOH) method for determination of Gram reactions of marine-bacteria. Appl Environ Microbiol 44:992–993. https://doi.org/10.0000/PMID6184019
Bernardet JF, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070. https://doi.org/10.1099/00207713-52-3-104
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for General and Molecular Bacteriology. American Society for Microbiology, Washington, pp 607–654
Komagata K, Suzuki KI (1987) Lipid and cell wall analysis in bacterial systematics. Methods Microbiol 19:161–207. https://doi.org/10.1016/S0580-9517(08)70410-0
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241. https://doi.org/10.1016/0167-7012(84)90018-6
Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2,4-diaminobutyric acid. J Appl Bacteriol 48:459–470. https://doi.org/10.1111/j.1365-2672.1980.tb01036.x
Coutinho PM, Deleury E, Davies GJ, Henrissat B (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328:307–317. https://doi.org/10.1016/S0022-2836(03)00307-3
Lairson LL, Henrissat B, Davies GJ, Withers SG (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem 77:521–555. https://doi.org/10.1146/annurev.biochem.76.061005.092322
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. https://doi.org/10.1099/ijs.0.64483-0
Konstantinidis KT, Tiedje JM (2005) Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A 102:2567–2572. https://doi.org/10.1073/pnas.0409727102
Funding
This work was supported by the Youth Foundation of the Shanxi Science and Technology Department (201901D211375), National Natural Science Foundation of China (32002143, 31972590), Excellent Talents Come to Shanxi to Reward Scientific Research Projects (SXYBKY2019024, SXYBKY2019025), Science and Technology Innovation Fund Project of Shanxi Agricultural University (2020BQ07), Animal Husbandry and ‘1331 project’ Key Discipline Construction program of Shanxi Province.
Author information
Authors and Affiliations
Contributions
CQX analyzed the data and wrote and revised the manuscript; HJN performed the experiments and wrote the manuscript; KSD, LG, and YJZ carried out chemotaxonomic analysis; LPS and QW performed phylogenetic analysis; YL and CXP designed and supervised the study. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This study was not performed on human or animals; therefore, no ethical approval is required.
Consent for Publication
All the authors agree to submit for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xia, CQ., Niu, HJ., Dong, KS. et al. Salinimicrobium sediminilitoris sp. nov., Isolated from a Tidal Flat. Curr Microbiol 79, 350 (2022). https://doi.org/10.1007/s00284-022-03037-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03037-7