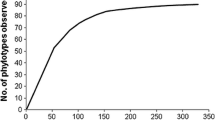
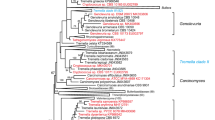
Abstract
Understanding diversity and distribution patterns of fungi, including yeasts, ultimately depends on accuracy of species recognition. However, different approaches to yeast species recognition often result in different entities or operational taxonomic units. We studied the effects of using different yeast species recognition approaches, namely morphological species recognition (MSR) and phylogenetic species recognition (PSR), on the distribution patterns of widespread basidiomycetous yeasts. Hence, we have revised a collection of yeast fungi isolated from spatially remote birch forests in the Moscow Region and Western Siberia with molecular typing and identification tools. PCR fingerprinting and rDNA sequencing analyses of strains of nine species previously identified on the basis of morphological and physiological tests (MSR) yielded 21 phylogenetic species (PSR), including three currently undescribed taxa. The number of distinct phylogenetic species comprised within a single morphospecies ranged from one to seven. A total of ten species were found in both regions, whereas the distribution of 11 yeasts was restricted to a single region only. Both geographical region and type of substrate (plant or soil) influence yeast distribution. Cryptococcus wieringae, C. victoriae, C. magnus, and Leucosporidium scottii were frequently found on plant substrates, whereas C. terricola and C. podzolicus were associated to soil substrates. Occurrence of C. magnus, C. albidus and Sporobolomyces roseus was found to depend on the geographical region. Microsatellite-PCR fingerprinting, MSP-PCR, applied to studying yeast intraspecific variability revealed three different types of distribution: (a) variability that depends on geographical factors (Curvibasidium cygneicollum, C. podzolicus, C. victoriae), (b) genetic identity irrespectively of the region of isolation (Rhodotorula pinicola, C. terricola), and (c) high degree of genetic variability that did not correlate with region of sampling (C. albidus and C. magnus).




Similar content being viewed by others
References
Agapow PM, Bininda-Emonds OR, Crandall KA, Gittleman JL, Mace GM, Marshall JC, Purvis A (2004) The impact of species concept on biodiversity studies. Q Rev Biol 79:161–179
Bab’eva IP, Chernov IY (1995) Geographical aspects of yeast ecology. Physiol Gen Biol Rev 9:1–54
Bab’eva IP, Reshetova IS (1975) A new yeast species Candida podzolica sp. nov. isolated from the soil. Microbiologia 44:333–338 English abstract
Barnett JA, Payne RW, Yarrow D (1990) Yeasts: characteristics and identification, 2nd edn. Cambridge University Press, Cambridge
Birkhofer K, Schöning I, Alt F, Herold N, Klarner B et al (2012) General relationships between abiotic soil properties and soil biota across spatial scales and different land-use types. PLoS ONE 7:e43292
Chernov IY (2005) Latitude-zonal and spatial-successional trends in yeast fungi distribution. Zh Obshch Biol 66:123–135 English abstract
Chernov IY, Bab’eva IP, Reshetova IS (1997) Synecology of yeast fungi in subtropical deserts. Usp Sovrem Biol 117:584–602 English abstract
Coelho MA, Gonçalves P, Sampaio JP (2011) Evidence for maintenance of sex determinants but not of sexual stages in red yeasts, a group of early diverged basidiomycetes. BMC Evol Biol 11:249
de Garcia V, Zalar P, Brizzio S, Gunde-Cimerman N, van Broock M (2012) Cryptococcus species (Tremellales) from glacial biomes in the southern (Patagonia) and northern (Svalbard) hemispheres. FEMS Microbiol Ecol 82:523–539
Dunlap CA, Evans KO, Theelen B, Boekhout T, Schisler DA (2007) Osmotic shock tolerance and membrane fluidity of cold-adapted Cryptococcus flavescens OH 182.9, previously reported as C. nodaensis, a biocontrol agent of Fusarium head blight. FEMS Yeast Res 7:449–458
Fonseca Á (1992) Utilization of tartaric acid and related compounds by yeasts: taxonomic implications. Can J Microbiol 38:1242–1251
Fonseca Á, Inácio J (2006) Phylloplane Yeasts. In: Peter G, Rosa C (eds) Biodiversity and Ecophysiology of Yeasts. Springer-Verlag, Berlin, pp 263–301
Fonseca Á, Scorzetti G, Fell JW (2000) Diversity in the yeast Cryptococcus albidus and related species as revealed by ribosomal DNA sequence analysis. Can J Microbiol 46:7–27
Fonseca Á, Boekhout T, Fell JW (2011) Cryptococcus Vuillemin. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, Amsterdam, pp 1661–1737
Gadanho M, Sampaio JP (2002) Polyphasic taxonomy of the basidiomycetous yeast genus Rhodotorula: Rh. glutinis sensu stricto and Rh. dairenensis comb. nov. FEMS Yeast Res 2:47–58
Ganter PF, Cardinali G, Giammaria M, Quarles B (2004) Correlations among measures of phenotypic and genetic variation within an oligotrophic asexual yeast, Candida sonorensis, collected from Opuntia. FEMS Yeast Res 4:527–540
Glushakova AM, Chernov IY (2004) Seasonal dynamics in a yeast population on leaves of the common wood sorrel Oxalis acetosella L. Microbiology 73:184–188
Glushakova AM, Zheltikova TM, Chernov IY (2004) Groups and sources of yeasts in house dust. Microbiology 73:94–98
Golubtsova YV, Glushakova AM, Chernov IY (2007) The seasonal dynamics of yeast communities in the rhizosphere of soddy-podzolic soils. Eurasian Soil Sci 40:875–879
Herzberg M, Fischer R, Titze A (2002) Conflicting results obtained by RAPD-PCR and large-subunit rDNA sequences in determining and comparing yeast strains isolated from flowers: a comparison of two methods. Int J Syst Evol Micr 52:1423–1433
Hong SG, Lee KH, Bae KS (2002) Diversity of yeasts associated with natural environments in Korea. Journal Microbiol 40:55–62
Inácio J, Rodrigues MG, Sobral P, Fonseca Á (2004) Characterisation and classification of phylloplane yeasts from Portugal related to the genus Taphrina and description of five novel Lalaria species. FEMS Yeast Res 4:541–555
Inácio J, Portugal L, Spencer-Martins I, Fonseca Á (2005) Phylloplane yeasts from Portugal: seven novel anamorphic species in the Tremellales lineage of the Hymenomycetes (Basidiomycota) producing orange-coloured colonies. FEMS Yeast Res 5:1167–1183
Kachalkin AV, Yurkov AM (2012) Yeast communities in Sphagnum phyllosphere along the temperature-moisture ecocline in the boreal forest-swamp ecosystem and description of Candida sphagnicola sp. nov. Anton Leeuw Int J G 102:29–43
Kachalkin AV, Glushakova AM, Yurkov AM, Chernov IY (2008) Characterization of yeast groupings in the phyllosphere of Sphagnum mosses. Microbiology 77:474–481
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform (describes the FFT-NS-1, FFT-NS-2 and FFT-NS-i strategies). Nucleic Acids Res 30:3059–3066
Kurtzman CP, Fell JW (1998) The yeasts, a taxonomic study, 4th edn. Elsevier, Amsterdam
Kurtzman CP, Fell JW (2006) Yeast systematics and phylogeny—implications of molecular identification methods for studies in ecology. In: Peter G, Rosa C (eds) Biodiversity and ecophysiology of yeasts. Springer-Verlag, Berlin, pp 11–30
Lachance M-A (2006) Yeast biodiversity: how many and how much. In: Peter G, Rosa C (eds) Biodiversity and ecophysiology of yeasts. Springer-Verlag, Berlin, pp 1–11
Lachance M-A, Starmer WT, Rosa CA, Bowles JM, Barker JSF, Janzen DH (2001) Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res 1:1–8
Lachance M-A, Dobson J, Wijayanayaka DN, Smith AM (2010) The use of parsimony network analysis for the formal delineation of phylogenetic species of yeasts: Candida apicola, Candida azyma, and Candida parazyma sp. nov., cosmopolitan yeasts associated with floricolous insects. Anton Leeuw Int J G 97:155–170
Leducq JB, Charron G, Samani P, Dubé AK, Sylvester K et al (2014) Local climatic adaptation in a widespread microorganism. P Roy Soc B-Biol Sci 281:20132472
Liti G, Carter DM, Moses AM, Warringer J, Parts L et al (2009) Population genomics of domestic and wild yeasts. Nature 458:337–341
Maksimova IA, Chernov IY (2004) Community structure of yeast fungi in forest biogeocenoses. Microbiology 73:474–481
Maksimova IA, Yurkov AM, Chernov IY (2009) Spatial structure of epiphytic yeast communities on fruits of Sorbus aucuparia L. Biol Bull 36:613–618
Naumov GI, Naumova ES, Molina FI (2000) Genetic variations among European strains of Saccharomyces paradoxus: results from DNA fingerprinting. Syst Appl Microbiol 23:86–92
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Sampaio JP (1999) Utilization of low molecular weight aromatic compounds by heterobasidiomycetous yeasts: taxonomic implications. Can J Microbiol 45:491–512
Sampaio JP (2011) Rhodotorula Harrison (1928). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, San Diego, pp 1873–1928
Sampaio JP (2011) Curvibasidium Sampaio and Golubev (2004). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, San Diego, pp 1413–1418
Sampaio JP (2011) Leucosporidium Fell, Statzell, Hunter & Phaff (1969). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, San Diego, pp 1489–1494
Sampaio JP, Gadanho M, Santos S, Duarte FL, Pais C, Fonseca Á, Fell JW (2001) Polyphasic taxonomy of the basidiomycetous yeast genus Rhodosporidium: R. kratochvilovae and related anamorphic species. Int J Syst Evol Micr 51:687–697
Sampaio JP, Golubev WI, Fell JW, Gadanho M, Golubev NW (2004) Curvibasidium cygneicollum gen. nov., sp. nov., Curvibasidium pallidicorallinum sp. nov., novel taxa in the Microbotryomycetidae, Urediniomycetes, and the relationship with Rhodotorula fujisanensis and Rhodotorula nothofagi. Int J Syst Evol Micr 54:1401–1407
Scorzetti G, Fell JW, Fonseca Á, Statzell-Tallman A (2002) Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res 2:495–517
Sites JW Jr, Crandall KA (1997) Testing species boundaries in biodiversity studies. Conserv Biol 11:1289–1297
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol 75:758–771
Starmer WT, Aberdeen V, Lachance M-A (2006) The biogeographic diversity of cactophilic yeasts. In: Peter G, Rosa C (eds) Biodiversity and ecophysiology of yeasts. Springer-Verlag, Berlin, pp 485–499
Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D (2006) Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philos T R Soc B 361:1947–1963
Thomas-Hall S, Watson K, Scorzetti G (2002) Cryptococcus statzelliae sp. nov. and three novel strains of Cryptococcus victoriae, yeasts isolated from Antarctic soils. Int J Syst Evol Micr 52:2303–2308
Vishniac HS (1995) Simulated in situ competitive ability and survival of a representative soil yeast, Cryptococcus albidus. Microb Ecol 30:309–320
Vishniac HS (2002) Cryptococcus tephrensis sp. nov., and Cryptococcus heimaeyensis sp. nov.; new anamorphic basidiomycetous yeast species from Iceland. Can J Microbiol 48:463–467
Vishniac HS (2006) A multivariate analysis of soil yeasts isolated from a latitudinal gradient. Microbl Ecol 52:90–103
Walsh PD (2000) Sample size for the diagnosis of conservation units. Conserv Biol 14:1533–1537
Wang QM, Boekhout T, Bai FY (2011) Cryptococcus foliicola sp. nov. and Cryptococcus taibaiensis sp. nov., novel basidiomycetous yeast species from plant leaves. J Gen Appl Microbiol 57:285–291
Whitaker RJ (2006) Allopatric origins of microbial species. Philos T R Soc B 361:1975–1984
Wuczkowski M, Prillinger H (2004) Molecular identification of yeasts from soils of the alluvial forest national park along the river Danube downstream of Vienna, Austria (“Nationalpark Donauauen”). Microbiol Res 159:263–275
Yarrow D (1998) Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. Elsevier, Amsterdam, pp 77–100
Yurkov AM, Chernov IY (2005) Geographical races of certain species of ascomycetous yeasts in the Moscow and Novosibirsk regions. Microbiology 74:597–601
Yurkov AM, Golubev WI (2013) Phylogenetic study of Cryptococcus laurentii mycocinogenic strains. Mycol Prog 12:777–782
Yurkov AM, Maksimova IA, Chernov IY (2004) The comparative analysis of yeast communities structure in birch forests of European Russia and Western Siberia. Mikol Fitopatol 38:71–79 English abstract
Yurkov AM, Kemler M, Begerow D (2011) Species accumulation curves and incidence-based species richness estimators to appraise the diversity of cultivable yeasts from beech forest soils. PLoS ONE 6:e23671
Yurkov AM, Kemler M, Begerow D (2012) Assessment of yeast diversity in soils under different management regimes. Fungal Ecol 5:24–35
Yurkov AM, Krüger D, Begerow D, Arnold N, Tarkka MT (2012) Basidiomycetous yeasts from Boletales fruiting bodies and their interactions with the mycoparasite Sepedonium chrysospermum and the host fungus Paxillus. Microb Ecol 63:295–303
Acknowledgments
AY received funds from the cooperation agreement “Acordo Cultural Portugal - Rússia” (GRICES), Russian Fund for Basic Research (RFBR 04-04-48713) and Fundação para a Ciência e a Tecnologia (Grants SFRH/BPD/79620/2011, PTDC/BIA-BIC/4585/2012). J.P. Sampaio and M. Gadanho (CREM, Portugal) are acknowledged for obtaining sequence data from two isolates. A.M. Glushakova (MSU, Moscow) and A.A. Sakharov assisted in samples collection.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yurkov, A., Inácio, J., Chernov, I.Y. et al. Yeast Biogeography and the Effects of Species Recognition Approaches: The Case Study of Widespread Basidiomycetous Species from Birch Forests in Russia. Curr Microbiol 70, 587–601 (2015). https://doi.org/10.1007/s00284-014-0755-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-014-0755-9