Abstract
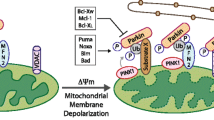
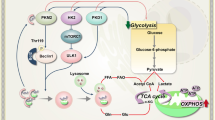
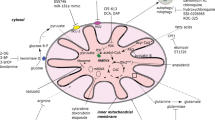
Mitophagy, the selective autophagic process that specifically degrades mitochondria, serves as a vital regulatory mechanism for eliminating damaged mitochondria and maintaining cellular balance. Emerging research underscores the central role of mitophagy in the initiation, advancement, and treatment of cancer. Mitophagy is widely acknowledged to govern mitochondrial homeostasis in hematopoietic stem cells (HSCs), influencing their metabolic dynamics. In this article, we integrate recent data to elucidate the regulatory mechanisms governing mitophagy and its intricate significance in the context of leukemia. An in-depth molecular elucidation of the processes governing mitophagy may serve as a basis for the development of pioneering approaches in targeted therapeutic interventions.



Similar content being viewed by others
Data availability
No datasets were generated or analyzed during the current study.
References
Song C, Pan S, Zhang J, Li N, Geng Q (2022) Mitophagy A. novel perspective for insighting into cancer and cancer treatment. Cell Prolif. 55(12):e13327. https://doi.org/10.1111/cpr.13327
Spinelli JB, Haigis MC (2018) The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol 20(7):745–754. https://doi.org/10.1038/s41556-018-0124-1
Bock FJ, Tait SWG (2020) Mitochondria as multifaceted regulators of cell death. Nat Rev Mol Cell Biol 21(2):85–100. https://doi.org/10.1038/s41580-019-0173-8
Oanh NTK, Lee H-S, Kim Y-H, Min S, Park Y-J, Heo J, Park Y-Y, Lim W-C, Cho H (2022) Regulation of nuclear DNA damage response by mitochondrial morphofunctional pathway. Nucleic Acids Res 50(16):9247–9259. https://doi.org/10.1093/nar/gkac690
Wang S, Long H, Hou L, Feng B, Ma Z, Wu Y, Zeng Y, Cai J, Zhang D-W, Zhao G (2023) The mitophagy pathway and its implications in human diseases. Signal Transduct Target Ther 8(1):304. https://doi.org/10.1038/s41392-023-01503-7
Lu Y, Li Z, Zhang S, Zhang T, Liu Y, Zhang L (2023) Cellular mitophagy Mechanism, roles in diseases and small molecule pharmacological regulation. Theranostics 13(2):736–766. https://doi.org/10.7150/thno.79876
Siegel RL, Miller KD, Wagle NS (2023) Jemal A (2023) Cancer statistics. CA Cancer J Clin 73(1):17–48. https://doi.org/10.3322/caac.21763
Onishi M, Yamano K, Sato M, Matsuda N, Okamoto K (2021) Molecular mechanisms and physiological functions of mitophagy. EMBO J 40(3):e10470. https://doi.org/10.15252/embj.2020104705
Titus AS, Sung E-A, Zablocki D, Sadoshima J (2023) Mitophagy for cardioprotection. Basic Res Cardiol 118(1):42. https://doi.org/10.1007/s00395-023-01009-x
Nguyen TN, Padman BS, Lazarou M (2016) Deciphering the molecular signals of PINK1/Parkin mitophagy. Trends Cell Biol 26(10):733–744. https://doi.org/10.1016/j.tcb.2016.05.008
Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SHY, Renton AEM, Harvey RJ, Whitworth AJ, Martins LM, Abramov AY, Wood NW (2011) PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet 20(5):867–879. https://doi.org/10.1093/hmg/ddq526
Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ (2010) Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 191(5):933–942. https://doi.org/10.1083/jcb.201008084
Greene AW, Grenier K, Aguileta MA, Muise S, Farazifard R, Haque ME, McBride HM, Park DS, Fon EA (2012) Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep 13(4):378–385. https://doi.org/10.1038/embor.2012.14
Shi G, McQuibban GA (2017) The mitochondrial rhomboid protease PARL is regulated by PDK2 to integrate mitochondrial quality control and metabolism. Cell Rep 18(6):1458–1472. https://doi.org/10.1016/j.celrep.2017.01.029
Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RLA, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrané J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S (2010) PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A 107(1):378–383. https://doi.org/10.1073/pnas.0911187107
Connelly EM, Frankel KS, Shaw GS (2023) Parkin and mitochondrial signalling. Cell Signal 106:110631. https://doi.org/10.1016/j.cellsig.2023.110631
Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL (2011) PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147(4):893–906. https://doi.org/10.1016/j.cell.2011.10.018
Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, Shaler T, Walker D, Yang Y, Regnstrom K, Diep L, Zhang Z, Chiou S, Bova M, Artis DR, Yao N, Baker J, Yednock T, Johnston JA (2013) Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun 4:1982. https://doi.org/10.1038/ncomms2982
McWilliams TG, Barini E, Pohjolan-Pirhonen R, Brooks SP, Singh F, Burel S, Balk K, Kumar A, Montava-Garriga L, Prescott AR, Hassoun SM, Mouton-Liger F, Ball G, Hills R, Knebel A, Ulusoy A, Di Monte DA, Tamjar J, Antico O, Fears K, Smith L, Brambilla R, Palin E, Valori M, Eerola-Rautio J, Tienari P, Corti O, Dunnett SB, Ganley IG, Suomalainen A, Muqit MMK (2018) Phosphorylation of Parkin at serine 65 is essential for its activation in vivo. Open Biol. 8(11):180108. https://doi.org/10.1098/rsob.180108
Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ (2015) The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524(7565):309–314. https://doi.org/10.1038/nature14893
Vargas JNS, Wang C, Bunker E, Hao L, Maric D, Schiavo G, Randow F, Youle RJ (2019) Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol Cell 74(2):347-362.e6. https://doi.org/10.1016/j.molcel.2019.02.010
Padman BS, Nguyen TN, Uoselis L, Skulsuppaisarn M, Nguyen LK, Lazarou M (2019) LC3/GABARAPs drive ubiquitin-independent recruitment of Optineurin and NDP52 to amplify mitophagy. Nat Commun 10(1):408. https://doi.org/10.1038/s41467-019-08335-6
Qiu Y, Wang J, Li H, Yang B, Wang J, He Q, Weng Q (2022) Emerging views of OPTN (optineurin) function in the autophagic process associated with disease. Autophagy 18(1):73–85. https://doi.org/10.1080/15548627.2021.1908722
Tao M, Liu T, You Q, Jiang Z (2020) p62 as a therapeutic target for tumor. Eur J Med Chem 193:112231. https://doi.org/10.1016/j.ejmech.2020.112231
Mukherjee R, Chakrabarti O (2016) Ubiquitin-mediated regulation of the E3 ligase GP78 by MGRN1 in trans affects mitochondrial homeostasis. J Cell Sci 129(4):757–773. https://doi.org/10.1242/jcs.176537
Fu L, Cui C-P, Zhang X, Zhang L (2020) The functions and regulation of Smurfs in cancers. Semin Cancer Biol 67(Pt 2):102–116. https://doi.org/10.1016/j.semcancer.2019.12.023
Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dötsch V, Ney PA, Dikic I (2010) Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep 11(1):45–51. https://doi.org/10.1038/embor.2009.256
He Y-L, Li J, Gong S-H, Cheng X, Zhao M, Cao Y, Zhao T, Zhao Y-Q, Fan M, Wu H-T, Zhu L-L, Wu L-Y (2022) BNIP3 phosphorylation by JNK1/2 promotes mitophagy via enhancing its stability under hypoxia. Cell Death Dis 13(11):966. https://doi.org/10.1038/s41419-022-05418-z
Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J (2008) Essential role for Nix in autophagic maturation of erythroid cells. Nature 454(7201):232–235. https://doi.org/10.1038/nature07006
Esteban-Martínez L, Sierra-Filardi E, McGreal RS, Salazar-Roa M, Mariño G, Seco E, Durand S, Enot D, Graña O, Malumbres M, Cvekl A, Cuervo AM, Kroemer G, Boya P (2017) Programmed mitophagy is essential for the glycolytic switch during cell differentiation. EMBO J 36(12):1688–1706. https://doi.org/10.15252/embj.201695916
Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson ÅB (2012) Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem 287(23):19094–19104. https://doi.org/10.1074/jbc.M111.322933
Zhang J, Loyd MR, Randall MS, Waddell MB, Kriwacki RW, Ney PA (2012) A short linear motif in BNIP3L (NIX) mediates mitochondrial clearance in reticulocytes. Autophagy 8(9):1325–1332. https://doi.org/10.4161/auto.20764
Shi R-Y, Zhu S-H, Li V, Gibson SB, Xu X-S, Kong J-M (2014) BNIP3 interacting with LC3 triggers excessive mitophagy in delayed neuronal death in stroke. CNS Neurosci Ther 20(12):1045–1055. https://doi.org/10.1111/cns.12325
Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL (2001) HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res 61(18):6669–6673
Cao Y, Zheng J, Wan H, Sun Y, Fu S, Liu S, He B, Cai G, Cao Y, Huang H, Li Q, Ma Y, Chen S, Wang F, Jiang H (2023) A mitochondrial SCF-FBXL4 ubiquitin E3 ligase complex degrades BNIP3 and NIX to restrain mitophagy and prevent mitochondrial disease. EMBO J 42(13):e113033. https://doi.org/10.15252/embj.2022113033
Zheng J, Cao Y, Yang J, Jiang H (2022) UBXD8 mediates mitochondria-associated degradation to restrain apoptosis and mitophagy. EMBO Rep 23(10):e54859. https://doi.org/10.15252/embr.202254859
Liu L, Feng Du, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, Huang L, Xue P, Li B, Wang X, Jin H, Wang J, Yang F, Liu P, Zhu Y, Sui S, Chen Q (2012) Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol 14(2):177–185. https://doi.org/10.1038/ncb2422
Chen Z, Liu L, Cheng Q, Li Y, Wu H, Zhang W, Wang Y, Sehgal SA, Siraj S, Wang X, Wang J, Zhu Y, Chen Q (2017) Mitochondrial E3 ligase MARCH5 regulates FUNDC1 to fine-tune hypoxic mitophagy. EMBO Rep 18(3):495–509. https://doi.org/10.15252/embr.201643309
Chen G, Han Z, Feng Du, Chen Y, Chen L, Wu H, Huang L, Zhou C, Cai X, Fu C, Duan L, Wang X, Liu L, Liu X, Shen Y, Zhu Y, Chen Q (2014) A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol Cell 54(3):362–377. https://doi.org/10.1016/j.molcel.2014.02.034
Murakawa T, Yamaguchi O, Hashimoto A, Hikoso S, Takeda T, Oka T, Yasui H, Ueda H, Akazawa Y, Nakayama H, Taneike M, Misaka T, Omiya S, Shah AM, Yamamoto A, Nishida K, Ohsumi Y, Okamoto K, Sakata Y, Otsu K (2015) Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat Commun 6:7527. https://doi.org/10.1038/ncomms8527
Lim GG, Lim K-L (2017) Parkin-independent mitophagy-FKBP8 takes the stage. EMBO Rep 18(6):864–865. https://doi.org/10.15252/embr.201744313
Wei Y, Chiang W-C, Sumpter R, Mishra P, Levine B (2017) Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell 168(1–2):224-238.e10. https://doi.org/10.1016/j.cell.2016.11.042
Xiao Y, Zhou Y, Lu Y, Zhou K, Cai W (2018) PHB2 interacts with LC3 and SQSTM1 is required for bile acids-induced mitophagy in cholestatic liver. Cell Death Dis 9(2):160. https://doi.org/10.1038/s41419-017-0228-8
Shen Z, Li Y, Gasparski AN, Abeliovich H, Greenberg ML (2017) Cardiolipin regulates mitophagy through the protein kinase C pathway. J Biol Chem 292(7):2916–2923. https://doi.org/10.1074/jbc.M116.753574
Rodriguez-Fraticelli AE, Wolock SL, Weinreb CS, Panero R, Patel SH, Jankovic M, Sun J, Calogero RA, Klein AM, Camargo FD (2018) Clonal analysis of lineage fate in native haematopoiesis. Nature 553(7687):212–216. https://doi.org/10.1038/nature25168
Morrison SJ, Wandycz AM, Hemmati HD, Wright DE, Weissman IL (1997) Identification of a lineage of multipotent hematopoietic progenitors. Development 124(10):1929–1939. https://doi.org/10.1242/dev.124.10.1929
Chen Z, Guo Q, Song G, Hou Y (2022) Molecular regulation of hematopoietic stem cell quiescence. Cell Mol Life Sci 79(4):218. https://doi.org/10.1007/s00018-022-04200-w
Xu B, Hu R, Liang Z, Chen T, Chen J, Hu Y, Jiang Y, Li Y (2021) Metabolic regulation of the bone marrow microenvironment in leukemia. Blood Rev 48:100786. https://doi.org/10.1016/j.blre.2020.100786
Koschade SE, Brandts CH (2020) Selective Autophagy in normal and malignant hematopoiesis. J Mol Biol 432(1):261–282. https://doi.org/10.1016/j.jmb.2019.06.025
Dzierzak E, Bigas A (2018) Blood development hematopoietic stem cell dependence and independence. Cell Stem Cell 22(5):639–651. https://doi.org/10.1016/j.stem.2018.04.015
Nakamura-Ishizu A, Takizawa H, Suda T (2014) The analysis, roles and regulation of quiescence in hematopoietic stem cells. Development 141(24):4656–4666. https://doi.org/10.1242/dev.106575
Kohli L, Passegué E (2014) Surviving change The metabolic journey of hematopoietic stem cells. Trends Cell Biol 24(8):479–487. https://doi.org/10.1016/j.tcb.2014.04.001
Wang Y-H, Israelsen WJ, Lee D, Yu VWC, Jeanson NT, Clish CB, Cantley LC, Vander Heiden MG, Scadden DT (2014) Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell 158(6):1309–1323. https://doi.org/10.1016/j.cell.2014.07.048
Ludin A, Gur-Cohen S, Golan K, Kaufmann KB, Itkin T, Medaglia C, Lu X-J, Ledergor G, Kollet O, Lapidot T (2014) Reactive oxygen species regulate hematopoietic stem cell self-renewal, migration and development, as well as their bone marrow microenvironment. Antioxid Redox Signal 21(11):1605–1619. https://doi.org/10.1089/ars.2014.5941
Jang Y-Y, Sharkis SJ (2007) A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood 110(8):3056–3063. https://doi.org/10.1182/blood-2007-05-087759
Takubo K, Nagamatsu G, Kobayashi CI, Nakamura-Ishizu A, Kobayashi H, Ikeda E, Goda N, Rahimi Y, Johnson RS, Soga T, Hirao A, Suematsu M, Suda T (2013) Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12(1):49–61. https://doi.org/10.1016/j.stem.2012.10.011
Jensen KS, Binderup T, Jensen KT, Therkelsen I, Borup R, Nilsson E, Multhaupt H, Bouchard C, Quistorff B, Kjaer A, Landberg G, Staller P (2011) FoxO3A promotes metabolic adaptation to hypoxia by antagonizing Myc function. EMBO J 30(22):4554–4570. https://doi.org/10.1038/emboj.2011.323
Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee C-H, Pandolfi PP (2012) A PML–PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med 18(9):1350–1358. https://doi.org/10.1038/nm.2882
Katajisto P, Döhla J, Chaffer CL, Pentinmikko N, Marjanovic N, Iqbal S, Zoncu R, Chen W, Weinberg RA, Sabatini DM (2015) Stem cells Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science 348(6232):340–343. https://doi.org/10.1126/science.1260384
Joffre C, Ducau C, Poillet-Perez L, Courdy C, Mansat-De Mas V (2021) Autophagy a Close relative of AML biology. Biology (Basel). 10(6):552. https://doi.org/10.3390/biology10060552
Murphy MP (2009) How mitochondria produce reactive oxygen species. Biochem J 417(1):1–13. https://doi.org/10.1042/BJ20081386
Ito K, Turcotte R, Cui J, Zimmerman SE, Pinho S, Mizoguchi T, Arai F, Runnels JM, Alt C, Teruya-Feldstein J, Mar JC, Singh R, Suda T, Lin CP, Frenette PS, Ito K (2016) Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science 354(6316):1156–1160. https://doi.org/10.1126/science.aaf5530
Morganti C, Ito K (2021) Mitochondrial contributions to hematopoietic stem cell aging. Int J Mol Sci. 22(20):11117. https://doi.org/10.3390/ijms222011117
Li J, Yang D, Li Z, Zhao M, Wang D, Sun Z, Wen P, Dai Y, Gou F, Ji Y, Zhao D, Yang L (2023) PINK1/Parkin-mediated mitophagy in neurodegenerative diseases. Ageing Res Rev 84:101817. https://doi.org/10.1016/j.arr.2022.101817
Vannini N, Girotra M, Naveiras O, Nikitin G, Campos V, Giger S, Roch A, Auwerx J, Lutolf MP (2016) Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nat Commun 7:13125. https://doi.org/10.1038/ncomms13125
Ho TT, Warr MR, Adelman ER, Lansinger OM, Flach J, Verovskaya EV, Figueroa ME, Passegué E (2017) Autophagy maintains the metabolism and function of young and old stem cells. Nature 543(7644):205–210. https://doi.org/10.1038/nature21388
Jin G, Xu C, Zhang X, Long J, Rezaeian AH, Liu C, Furth ME, Kridel S, Pasche B, Bian X-W, Lin H-K (2018) Atad3a suppresses Pink1-dependent mitophagy to maintain homeostasis of hematopoietic progenitor cells. Nat Immunol 19(1):29–40. https://doi.org/10.1038/s41590-017-0002-1
Albadari N, Deng S, Li W (2019) The transcriptional factors HIF-1 and HIF-2 and their novel inhibitors in cancer therapy. Expert Opin Drug Discov 14(7):667–682. https://doi.org/10.1080/17460441.2019.1613370
Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL (2007) HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 11(5):407–420. https://doi.org/10.1016/j.ccr.2007.04.001
Wu H, Chen Q (2015) Hypoxia activation of mitophagy and its role in disease pathogenesis. Antioxid Redox Signal 22(12):1032–1046. https://doi.org/10.1089/ars.2014.6204
Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouysségur J, Mazure NM (2009) Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 29(10):2570–2581. https://doi.org/10.1128/MCB.00166-09
Kocabas F, Zheng J, Thet S, Copeland NG, Jenkins NA, DeBerardinis RJ, Zhang C, Sadek HA (2012) Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood 120(25):4963–4972. https://doi.org/10.1182/blood-2012-05-432260
Rouault-Pierre K, Lopez-Onieva L, Foster K, Anjos-Afonso F, Lamrissi-Garcia I, Serrano-Sanchez M, Mitter R, Ivanovic Z, de Verneuil H, Gribben J, Taussig D, Rezvani HR, Mazurier F, Bonnet D (2013) HIF-2α protects human hematopoietic stem/progenitors and acute myeloid leukemic cells from apoptosis induced by endoplasmic reticulum stress. Cell Stem Cell 13(5):549–563. https://doi.org/10.1016/j.stem.2013.08.011
Vukovic M, Sepulveda C, Subramani C, Guitart AV, Mohr J, Allen L, Panagopoulou TI, Paris J, Lawson H, Villacreces A, Armesilla-Diaz A, Gezer D, Holyoake TL, Ratcliffe PJ, Kranc KR (2016) Adult hematopoietic stem cells lacking Hif-1α self-renew normally. Blood 127(23):2841–2846. https://doi.org/10.1182/blood-2015-10-677138
Bonnet D, Dick JE (1997) Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3(7):730–737. https://doi.org/10.1038/nm0797-730
Stergiou IE, Kapsogeorgou EK (2021) Autophagy and metabolism in normal and malignant hematopoiesis. Int J Mol Sci. 22(16):8540. https://doi.org/10.3390/ijms22168540
Liang H, Dong S, Fu W, Zhang S, Yu W, Dong F, He B, Wang J, Gao Y, Zhou Y, Ru Y (2022) Deciphering the heterogeneity of mitochondrial functions during hematopoietic lineage differentiation. Stem Cell Rev Rep 18(6):2179–2194. https://doi.org/10.1007/s12015-022-10354-8
Pei S, Minhajuddin M, Adane B, Khan N, Stevens BM, Mack SC, Lai S, Rich JN, Inguva A, Shannon KM, Kim H, Tan A-C, Myers JR, Ashton JM, Neff T, Pollyea DA, Smith CA, Jordan CT (2018) AMPK/FIS1-mediated mitophagy is required for self-renewal of human AML stem cells. Cell Stem Cell 23(1):86-100.e6. https://doi.org/10.1016/j.stem.2018.05.021
Meyer LM, Koschade SE, Vischedyk JB, Thoelken M, Gubas A, Wegner M, Basoglu M, Knapp S, Kaulich M, Eimer S, Shaid S, Brandts CH (2023) Deciphering the mitophagy receptor network identifies a crucial role for OPTN (optineurin) in acute myeloid leukemia. Autophagy. 19(11):2982–2996. https://doi.org/10.1080/15548627.2023.2230839
Lazarini M, Machado-Neto JA, Duarte AdSS, Pericole FV, Vieira KP, Niemann FS, Alvarez M, Traina F, Saad STO (2016) BNIP3L in myelodysplastic syndromes and acute myeloid leukemia Impact on disease outcome and cellular response to decitabine. Haematologica 101(11):e44–e44. https://doi.org/10.3324/haematol.2016.142521
Maggi F, Morelli MB, Tomassoni D, Marinelli O, Aguzzi C, Zeppa L, Nabissi M, Santoni G, Amantini C (2022) The effects of cannabidiol via TRPV2 channel in chronic myeloid leukemia cells and its combination with imatinib. Cancer Sci 113(4):1235–1249. https://doi.org/10.1111/cas.15257
Rothe K, Porter V, Jiang X (2019) Current outlook on autophagy in human leukemia. Foe in cancer stem cells and drug resistance, friend in new therapeutic interventions. Int J Mol Sci. 20(3):461. https://doi.org/10.3390/ijms20030461
Vara-Perez M, Felipe-Abrio B, Agostinis P (2019) Mitophagy in cancer. A tale of adaptation. Cells 8(5). https://doi.org/10.3390/cells8050493
Dany M, Gencer S, Nganga R, Thomas RJ, Oleinik N, Baron KD, Szulc ZM, Ruvolo P, Kornblau S, Andreeff M, Ogretmen B (2016) Targeting FLT3-ITD signaling mediates ceramide-dependent mitophagy and attenuates drug resistance in AML. Blood 128(15):1944–1958. https://doi.org/10.1182/blood-2016-04-708750
Morad SAF, MacDougall MR, Abdelmageed N, Kao L-P, Feith DJ, Tan S-F, Kester M, Loughran TP, Wang HG, Cabot MC (2019) Pivotal role of mitophagy in response of acute myelogenous leukemia to a ceramide-tamoxifen-containing drug regimen. Exp Cell Res 381(2):256–264. https://doi.org/10.1016/j.yexcr.2019.05.021
Rodrigo R, Mendis N, Ibrahim M, Ma C, Kreinin E, Roma A, Berg S, Blay J, Spagnuolo PA (2019) Knockdown of BNIP3L or SQSTM1 alters cellular response to mitochondria target drugs. Autophagy 15(5):900–907. https://doi.org/10.1080/15548627.2018.1558002
Hao B-B, Li X-J, Jia X-L, Wang Y-X, Zhai L-H, Li D-Z, Liu J, Zhang D, Chen Y-L, Xu Y-H, Lee S-K, Xu G-F, Chen X-H, Dang Y-J, Liu B, Tan M-J (2020) The novel cereblon modulator CC-885 inhibits mitophagy via selective degradation of BNIP3L. Acta Pharmacol Sin 41(9):1246–1254. https://doi.org/10.1038/s41401-020-0367-9
Ianniciello A, Zarou MM, Rattigan KM, Scott M, Dawson A, Dunn K, Brabcova Z, Kalkman ER, Nixon C, Michie AM, Copland M, Vetrie D, Ambler M, Saxty B, Helgason GV (2021) ULK1 inhibition promotes oxidative stress-induced differentiation and sensitizes leukemic stem cells to targeted therapy. Sci Transl Med. 13(613):eabd5016. https://doi.org/10.1126/scitranslmed.abd5016
Olivas-Aguirre M, Pérez-Chávez J, Torres-López L, Hernández-Cruz A, Pottosin I, Dobrovinskaya O (2023) Dexamethasone-induced fatty acid oxidation and autophagy/mitophagy are essential for T-all glucocorticoid resistance. Cancers (Basel). 15(2):445. https://doi.org/10.3390/cancers15020445
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, Y., Ma, Z. Leukemia and mitophagy: a novel perspective for understanding oncogenesis and resistance. Ann Hematol (2024). https://doi.org/10.1007/s00277-024-05635-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00277-024-05635-w