Abstract
Background
Cutaneous melanoma (CM) has long been recognized as a lethal form of cancer. Despite persistent research endeavors, the precise underlying pathological mechanisms remain largely unclear, and the optimal treatment for this patient population remains undetermined.
Objectives
This study aims to examine the causal associations between CM and 486 metabolites.
Methods
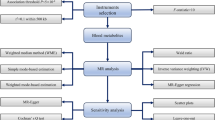
A two-sample Mendelian randomization (MR) analysis was conducted to ascertain the causal relationship between blood metabolites and CM. The causality analysis involved the inverse variance weighted (IVW) method, followed by the MR-Egger and weighted median (WM) methods. To increase the robustness of our findings, several sensitivity analyses, including the MR-Egger intercept, Cochran's Q test, and MR-pleiotropy residual sum and outlier (MR-PRESSO), were performed. The robustness of our results was further validated in independent outcome samples followed by a meta-analysis. Additionally, a metabolic pathway analysis was carried out.
Results
The two-sample MR analysis yielded a total of 27 metabolites as potential causal metabolites. After incorporating the outcomes of the sensitivity analyses, seven causal metabolites remained. Palmitoylcarnitine (OR 0.9903 95% CI 0.9848–0.9958, p = 0.0005) emerged as the sole metabolite with a significant causality after Bonferroni correction. Furthermore, the reverse MR analysis provided no evidence of reverse causality from CM to the identified metabolites.
Conclusions
This study suggested a causal relationship between seven human blood metabolites and the development of CM, thereby offering novel insights into the underlying mechanisms involved.
No Level Assigned
This journal requires that authors assign a level of evidence to each submission to which Evidence-Based Medicine rankings are applicable. This excludes Review Articles, Book Reviews, and manuscripts that concern Basic Science, Animal Studies, Cadaver Studies, and Experimental Studies. For a full description of these Evidence-Based Medicine ratings, please refer to the Table of Contents or the online Instructions to Authors www.springer.com/00266.



Similar content being viewed by others
Reference
Sauter ER, Herlyn M (1998) Molecular biology of human melanoma development and progression. Mol Carcinog 23(3):132–43
Slominski A, Wortsman J, Nickoloff B, McClatchey K, Mihm MC, Ross JS (1998) Molecular pathology of malignant melanoma. Am J Clin Pathol 110(6):788–794
Rastrelli M, Tropea S, Rossi CR, Alaibac M (2014) Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo 28(6):1005–11
Ahmed B, Qadir MI, Ghafoor S (2020) Malignant melanoma: skin cancer-diagnosis, prevention, and treatment. Crit Rev Eukaryot Gene Expr 30(4):291–297
Wang S, Chen Y, Sun J, Mo R, Tan Q (2023) Development and validation of two online dynamic nomograms for patients with non-distant metastatic cutaneous melanoma based on surgical approaches. Cancer Med 12(18):18479–90
Schmidt DR, Patel R, Kirsch DG, Lewis CA, Vander Heiden MG, Locasale JW (2021) Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J Clin 71(4):333–58
Liang L, Sun F, Wang H, Hu Z (2021) Metabolomics, metabolic flux analysis and cancer pharmacology. Pharmacol Ther 224:107827
Kosmopoulou M, Giannopoulou AF, Iliou A, Benaki D, Panagiotakis A, Velentzas AD et al (2020) Human melanoma-cell metabolic profiling: identification of novel biomarkers indicating metastasis. Int J Mol Sci 21(7):2436
Weber DD, Thapa M, Aminzadeh-Gohari S, Redtenbacher AS, Catalano L, Feichtinger RG et al (2021) Targeted metabolomics identifies plasma biomarkers in mice with metabolically heterogeneous melanoma xenografts. Cancers 13(3):434
Hagyousif YA, Sharaf BM, Zenati RA, El-Huneidi W, Bustanji Y, Abu-Gharbieh E et al (2023) Skin cancer metabolic profile assessed by different analytical platforms. Int J Mol Sci 24(2):1604
Johnson CH, Ivanisevic J, Siuzdak G (2016) Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol 17(7):451–9
Taylor NJ, Gaynanova I, Eschrich SA, Welsh EA, Garrett TJ, Beecher C et al (2020) Metabolomics of primary cutaneous melanoma and matched adjacent extratumoral microenvironment. PLoS One 15(10):e0240849
Liu Y, Cao Y, Zhang W, Bergmeier S, Qian Y, Akbar H et al (2012) A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol Cancer Ther 11(8):1672–82
Cao X, Fang L, Gibbs S, Huang Y, Dai Z, Wen P et al (2007) Glucose uptake inhibitor sensitizes cancer cells to daunorubicin and overcomes drug resistance in hypoxia. Cancer Chemother Pharmacol 59(4):495–505
Bennett DA (2010) An introduction to instrumental variables–part 2: Mendelian randomisation. Neuroepidemiology 35(4):307–310
Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G (2008) Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 27(8):1133–63
Elliott J, Bodinier B, Bond TA, Chadeau-Hyam M, Evangelou E, Moons KGM et al (2020) Predictive accuracy of a polygenic risk score-enhanced prediction model versus a clinical risk score for coronary artery disease. JAMA 323(7):636–645
Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J et al (2014) An atlas of genetic influences on human blood metabolites. Nat Genet 46(6):543–50
Burgess S, Thompson SG, Collaboration CCG (2011) Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol 40(3):755–64
Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan N, Thompson J (2017) A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med 36(11):1783–802
Burgess S, Thompson SG (2017) Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol 32(5):377–89
Bowden J, Davey Smith G, Haycock PC, Burgess S (2016) Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 40(4):304–314
Verbanck M, Chen CY, Neale B, Do R (2018) Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 50(5):693–8
Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40(Database issue):D109-14
Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G et al (2018) MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 46(W1):W486–W94
Vescovi G, Weber B, Matrat M, Ramacci C, Nabet P, Kremer B (1988) Modulation by palmitoyl-carnitine of calcium activated, phospholipid-dependent protein kinase activity and inhibition of melanoma cell growth. Br J Dermatol 119(2):171–178
Sumantran VN, Mishra P, Sudhakar N (2015) Microarray analysis of differentially expressed genes regulating lipid metabolism during melanoma progression. Indian J Biochem Biophys 52(2):125–31
Yan C, Wu D, Gan L, Wang J, Yang W, Xu B (2022) Significant metabolic alterations in non-small cell lung cancer patients by epidermal growth factor receptor-targeted therapy and PD-1/PD-L1 immunotherapy. Front Pharmacol 13:949745
Lin Z, Liu F, Shi P, Song A, Huang Z, Zou D et al (2018) Fatty acid oxidation promotes reprogramming by enhancing oxidative phosphorylation and inhibiting protein kinase C. Stem Cell Res Ther 9(1):47
Dong R, Ye N, Zhao S, Wang G, Zhang Y, Wang T et al (2021) Studies on novel diagnostic and predictive biomarkers of intrahepatic cholestasis of pregnancy through metabolomics and proteomics. Front Immunol 12:733225
Bouchouirab FZ, Fortin M, Noll C, Dube J, Carpentier AC (2018) Plasma palmitoyl-carnitine (AC16:0) is a marker of increased postprandial nonesterified incomplete fatty acid oxidation rate in adults with type 2 diabetes. Can J Diabetes 42(4):382–8
Wenzel U, Nickel A, Daniel H (2005) Increased carnitine-dependent fatty acid uptake into mitochondria of human colon cancer cells induces apoptosis. J Nutr 135(6):1510–4
Turnbull PC, Hughes MC, Perry CGR (2019) The fatty acid derivative palmitoylcarnitine abrogates colorectal cancer cell survival by depleting glutathione. Am J Physiol Cell Physiol 317(6):C1278–C1288
Beloribi-Djefaflia S, Vasseur S, Guillaumond F (2016) Lipid metabolic reprogramming in cancer cells. Oncogenesis 5(1):e189
Fu Y, Rathod D, Patel K (2020) Protein kinase C inhibitor anchored BRD4 PROTAC PEGylated nanoliposomes for the treatment of vemurafenib-resistant melanoma. Exp Cell Res 396(1):112275
Sobiesiak-Mirska J, Nałecz MJ, Nałecz KA (2003) Interaction of palmitoylcarnitine with protein kinase C in neuroblastoma NB-2a cells. Neurochem Int 42(1):45–55
Alberg AJ, Gordon GB, Hoffman SC, Comstock GW, Helzlsouer KJ (2000) Serum dehydroepiandrosterone and dehydroepiandrosterone sulfate and the subsequent risk of developing colon cancer. Cancer Epidemiol Biomark. Prev. 9(5):517–21
Alberg AJ, Gordon GB, Genkinger JM, Hoffman SC, Selvin E, Comstock GW et al (2001) Serum dehydroepiandrosterone and dehydroepiandrosterone sulfate and risk of melanoma or squamous cell carcinoma of the skin. Anticancer Res 21(6a):4051–4
Yang P, Cartwright CA, Li J, Wen S, Prokhorova IN, Shureiqi I et al (2012) Arachidonic acid metabolism in human prostate cancer. Int J Oncol 41(4):1495–503
Chang J, Jiang L, Wang Y, Yao B, Yang S, Zhang B et al (2015) 12/15 Lipoxygenase regulation of colorectal tumorigenesis is determined by the relative tumor levels of its metabolite 12-HETE and 13-HODE in animal models. Oncotarget 6(5):2879–2888
Liu Q, Tan W, Che J, Yuan D, Zhang L, Sun Y et al (2018) 12-HETE facilitates cell survival by activating the integrin-linked kinase/NF-kappaB pathway in ovarian cancer. Cancer Manag Res 10:5825–38
Winer I, Normolle DP, Shureiqi I, Sondak VK, Johnson T, Su L et al (2002) Expression of 12-lipoxygenase as a biomarker for melanoma carcinogenesis. Melanoma Res 12(5):429–34
Honn KV, Tang DG, Gao X, Butovich IA, Liu B, Timar J et al (1994) 12-lipoxygenases and 12(S)-HETE: role in cancer metastasis. Cancer Metastasis Rev 13(3–4):365–96
Kang KH, Ling TY, Liou HH, Huang YK, Hour MJ, Liou HC et al (2013) Enhancement role of host 12/15-lipoxygenase in melanoma progression. Eur J Cancer. 49(12):2747–59
Long GV, Swetter SM, Menzies AM, Gershenwald JE, Scolyer RA (2023) Cutaneous melanoma. Lancet 402(10400):485–502
Rashid S, Shaughnessy M, Tsao H (2023) Melanoma classification and management in the era of molecular medicine. Dermatol Clin 41(1):49–63
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest to disclose.
Human and Animal Rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
For this type of study, informed consent is not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, X., Gao, Y., Fu, L. et al. Cutaneous Melanoma and 486 Human Blood Metabolites: A Mendelian Randomization Study. Aesth Plast Surg (2024). https://doi.org/10.1007/s00266-024-03873-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00266-024-03873-x