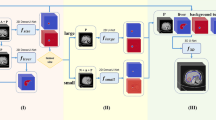
Abstract
Purpose
Liver Imaging Reporting and Data System (LI-RADS) is limited by interreader variability. Thus, our study aimed to develop a deep-learning model for classifying LI-RADS major features using subtraction images using magnetic resonance imaging (MRI).
Methods
This single-center retrospective study included 222 consecutive patients who underwent resection for hepatocellular carcinoma (HCC) between January, 2015 and December, 2017. Subtraction arterial, portal venous, and transitional phase images of preoperative gadoxetic acid-enhanced MRI were used to train and test the deep-learning models. Initially, a three-dimensional (3D) nnU-Net-based deep-learning model was developed for HCC segmentation. Subsequently, a 3D U-Net-based deep-learning model was developed to assess three LI-RADS major features (nonrim arterial phase hyperenhancement [APHE], nonperipheral washout, and enhancing capsule [EC]), utilizing the results determined by board-certified radiologists as reference standards. The HCC segmentation performance was assessed using the Dice similarity coefficient (DSC), sensitivity, and precision. The sensitivity, specificity, and accuracy of the deep-learning model for classifying LI-RADS major features were calculated.
Results
The average DSC, sensitivity, and precision of our model for HCC segmentation were 0.884, 0.891, and 0.887, respectively, across all the phases. Our model demonstrated a sensitivity, specificity, and accuracy of 96.6% (28/29), 66.7% (4/6), and 91.4% (32/35), respectively, for nonrim APHE; 95.0% (19/20), 50.0% (4/8), and 82.1% (23/28), respectively, for nonperipheral washout; and 86.7% (26/30), 54.2% (13/24), and 72.2% (39/54) for EC, respectively.
Conclusion
We developed an end-to-end deep-learning model that classifies the LI-RADS major features using subtraction MRI images. Our model exhibited satisfactory performance in classifying LI-RADS major features.
Graphical Abstract






Similar content being viewed by others
References
Chernyak V, Fowler KJ, Kamaya A, Kielar AZ, Elsayes KM, Bashir MR, Kono Y, Do RK, Mitchell DG, Singal AG (2018) Liver Imaging Reporting and Data System (LI-RADS) version 2018: imaging of hepatocellular carcinoma in at-risk patients. Radiology 289 (3):816. https://doi.org/10.1148/radiol.2018181494
Lee SM, Lee JM, Ahn SJ, Kang H-J, Yang HK, Yoon JH (2019) LI-RADS version 2017 versus version 2018: diagnosis of hepatocellular carcinoma on gadoxetate disodium–enhanced MRI. Radiology 292 (3):655-663. https://doi.org/10.1148/radiol.2019182867
Lee S, Kim SS, Roh YH, Choi JY, Park MS, Kim MJ (2020) Diagnostic performance of CT/MRI liver imaging reporting and data system v2017 for hepatocellular carcinoma: a systematic review and meta‐analysis. Liver Int 40 (6):1488-1497. https://doi.org/10.1111/liv.14424
Park J, Lee JM, Kim T-H, Yoon JH (2022) Imaging diagnosis of hepatocellular carcinoma: Future directions with special emphasis on hepatobiliary magnetic resonance imaging and contrast-enhanced ultrasound. Clin Mol Hepatol 28 (3):362. https://doi.org/10.3350/cmh.2021.0361
Fowler KJ, Tang A, Santillan C, Bhargavan-Chatfield M, Heiken J, Jha RC, Weinreb J, Hussain H, Mitchell DG, Bashir MR (2018) Interreader reliability of LI-RADS version 2014 algorithm and imaging features for diagnosis of hepatocellular carcinoma: a large international multireader study. Radiology 286 (1):173-185. https://doi.org/10.1148/radiol.2017170376
Barth BK, Donati OF, Fischer MA, Ulbrich EJ, Karlo CA, Becker A, Seifert B, Reiner CS (2016) Reliability, validity, and reader acceptance of LI-RADS—an in-depth analysis. Acad Radiol 23 (9):1145-1153. https://doi.org/10.1016/j.acra.2016.03.014
Ehman EC, Behr SC, Umetsu SE, Fidelman N, Yeh BM, Ferrell LD, Hope TA (2016) Rate of observation and inter-observer agreement for LI-RADS major features at CT and MRI in 184 pathology proven hepatocellular carcinomas. Abdom Radiol 41 (5):963-969. https://doi.org/10.1007/s00261-015-0623-5
Schellhaas B, Hammon M, Strobel D, Pfeifer L, Kielisch C, Goertz RS, Cavallaro A, Janka R, Neurath MF, Uder M (2018) Interobserver and intermodality agreement of standardized algorithms for non-invasive diagnosis of hepatocellular carcinoma in high-risk patients: CEUS-LI-RADS versus MRI-LI-RADS. Eur radiol 28 (10):4254-4264. https://doi.org/10.1007/s00330-018-5379-1
Wataya T, Yanagawa M, Tsubamoto M, Sato T, Nishigaki D, Kita K, Yamagata K, Suzuki Y, Hata A, Kido S (2023) Radiologists with and without deep learning–based computer-aided diagnosis: comparison of performance and interobserver agreement for characterizing and diagnosing pulmonary nodules/masses. Eur Radiol 33 (1):348-359. https://doi.org/10.1007/s00330-022-08948-4
Lim DSW, Makmur A, Zhu L, Zhang W, Cheng AJ, Sia DSY, Eide SE, Ong HY, Jagmohan P, Tan WC (2022) Improved productivity using deep learning–assisted reporting for lumbar spine MRI. Radiology 305 (1):160-166. https://doi.org/10.1148/radiol.220076
Wang M, Fu F, Zheng B, Bai Y, Wu Q, Wu J, Sun L, Liu Q, Liu M, Yang Y (2021) Development of an AI system for accurately diagnose hepatocellular carcinoma from computed tomography imaging data. Br J Cancer 125 (8):1111-1121. https://doi.org/10.1038/s41416-021-01511-w
Yasaka K, Akai H, Abe O, Kiryu S (2018) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 286 (3):887-896. https://doi.org/10.1148/radiol.2017170706
Hamm CA, Wang CJ, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin M, Duncan JS, Weinreb JC, Chapiro J (2019) Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol 29 (7):3338-3347. https://doi.org/10.1007/s00330-019-06205-9
Wu Y, White GM, Cornelius T, Gowdar I, Ansari MH, Supanich MP, Deng J (2020) Deep learning LI-RADS grading system based on contrast enhanced multiphase MRI for differentiation between LR-3 and LR-4/LR-5 liver tumors. Ann Transl Med 8(11):701. https://doi.org/10.21037/atm.2019.12.151
Wang CJ, Hamm CA, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin M, Weinreb JC, Duncan JS, Chapiro J (2019) Deep learning for liver tumor diagnosis part II: convolutional neural network interpretation using radiologic imaging features. Eur Radiol 29 (7):3348-3357. https://doi.org/10.1007/s00330-019-06214-8
Liang D, Lin L, Hu H, Zhang Q, Chen Q, Han X, Chen Y-W (2018) Combining convolutional and recurrent neural networks for classification of focal liver lesions in multi-phase CT images. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11071. Springer, Cham, pp 666-675. https://doi.org/10.1007/978-3-030-00934-2_74
Lee S-G, Kim E, Bae JS, Kim JH, Yoon S (2023) Robust End-to-End Focal Liver Lesion Detection Using Unregistered Multiphase Computed Tomography Images. IEEE Transactions on Emerging Topics in Computational Intelligence 7 (2):319-329. https://doi.org/10.1109/TETCI.2021.3132382
Isensee F, Jaeger PF, Kohl SA, Petersen J, Maier-Hein KH (2021) nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18 (2):203-211. https://doi.org/10.1038/s41592-020-01008-z
Ronneberger O, Fischer P, Brox T (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab, N., Hornegger, J., Wells, W., Frangi, A. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science(), vol 9351. Springer, Cham, pp 234-241. https://doi.org/10.1007/978-3-319-24574-4_28
Funding
This study was supported by grant No. ‘04-2020-2280’ from the Seoul National University Hospital Research Fund.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Jong-Min Kim and Joseph Nathanael Witanto are the employees of MEDICAL IP Co. Ltd. Sang Joon Park is the founder and CEO of MEDICAL IP Co. Ltd. Other authors have no competing interests related to this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Park, J., Bae, J.S., Kim, JM. et al. Development of a deep-learning model for classification of LI-RADS major features by using subtraction images of MRI: a preliminary study. Abdom Radiol 48, 2547–2556 (2023). https://doi.org/10.1007/s00261-023-03962-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-023-03962-6