Abstract
Purpose
The purpose of the study was to propose a deep transfer learning (DTL)-based model to distinguish indolent from clinically significant prostate cancer (PCa) lesions and to compare the DTL-based model with a deep learning (DL) model without transfer learning and PIRADS v2 score on 3 Tesla multi-parametric MRI (3T mp-MRI) with whole-mount histopathology (WMHP) validation.
Methods
With IRB approval, 140 patients with 3T mp-MRI and WMHP comprised the study cohort. The DTL-based model was trained on 169 lesions in 110 arbitrarily selected patients and tested on the remaining 47 lesions in 30 patients. We compared the DTL-based model with the same DL model architecture trained from scratch and the classification based on PIRADS v2 score with a threshold of 4 using accuracy, sensitivity, specificity, and area under curve (AUC). Bootstrapping with 2000 resamples was performed to estimate the 95% confidence interval (CI) for AUC.
Results
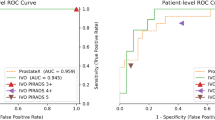
After training on 169 lesions in 110 patients, the AUC of discriminating indolent from clinically significant PCa lesions of the DTL-based model, DL model without transfer learning and PIRADS v2 score ≥ 4 were 0.726 (CI [0.575, 0.876]), 0.687 (CI [0.532, 0.843]), and 0.711 (CI [0.575, 0.847]), respectively, in the testing set. The DTL-based model achieved higher AUC compared to the DL model without transfer learning and PIRADS v2 score ≥ 4 in discriminating clinically significant lesions in the testing set.
Conclusion
The DeLong test indicated that the DTL-based model achieved comparable AUC compared to the classification based on PIRADS v2 score (p = 0.89).





Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68:7–30.
Ahmed HU, Akin O, Coleman JA, Crane S, Emberton M, Goldenberg L, Hricak H, Kattan MW, Kurhanewicz J, Moore CM, Parker C, Polascik TJ, Scardino P, van As N, Villers A (2012) Transatlantic Consensus Group on active surveillance and focal therapy for prostate cancer. BJU Int 109:1636–1647.
Lavery HJ, Droller MJ (2012) Do gleason patterns 3 and 4 prostate cancer represent separate disease states? J Urol 188:1667–1675.
Felker ER, Raman SS, Margolis DJ, Lu DSK, Shaheen N, Natarajan S, Sharma D, Huang J, Dorey F, Marks LS (2017) Risk Stratification Among Men With Prostate Imaging Reporting and Data System version 2 Category 3 Transition Zone Lesions: Is Biopsy Always Necessary? Am J Roentgenol 209:1272–1277.
Schröder FH, Hugosson J, Roobol MJ, Tammela TLJ, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, Denis LJ, Recker F, Berenguer A, Määttänen L, Bangma CH, Aus G, Villers A, Rebillard X, van der Kwast T, Blijenberg BG, Moss SM, de Koning HJ, Auvinen A (2009) Screening and Prostate-Cancer Mortality in a Randomized European Study. N Engl J Med 360:1320–1328.
Caster JM, Falchook AD, Hendrix LH, Chen RC (2015) Risk of Pathologic Upgrading or Locally Advanced Disease in Early Prostate Cancer Patients Based on Biopsy Gleason Score and PSA: A Population-Based Study of Modern Patients. Int J Radiat Oncol Biol Phys 92:244–51.
Cohen MS, Hanley RS, Kurteva T, Ruthazer R, Silverman ML, Sorcini A, Hamawy K, Roth RA, Tuerk I, Libertino JA (2008) Comparing the Gleason Prostate Biopsy and Gleason Prostatectomy Grading System: The Lahey Clinic Medical Center Experience and an International Meta-Analysis. Eur Urol 54:371–381.
Hoeks CMA, Barentsz JO, Hambrock T, Yakar D, Somford DM, Heijmink SWTPJ, Scheenen TWJ, Vos PC, Huisman H, van Oort IM, Witjes JA, Heerschap A, Fütterer JJ (2011) Prostate cancer: multiparametric MR imaging for detection, localization, and staging. Radiology 261:46–66.
Tan N, Margolis DJ, Lu DY, King KG, Huang J, Reiter RE, Raman SS (2015) Characteristics of detected and missed prostate cancer foci on 3-T multiparametric MRI using an endorectal coil correlated with whole-mount thin-section histopathology. Am J Roentgenol 205:W87–W92.
Litjens GJS, Barentsz JO, Karssemeijer N, Huisman HJ (2015) Clinical evaluation of a computer-aided diagnosis system for determining cancer aggressiveness in prostate MRI. Eur Radiol 3187–3199.
ACM (2015) Pi-Rads Prostate Imaging - Reporting and Data System. Am. Coll. Radiol.
Giannarini G, Girometti R, Sioletic S, Rossanese M, Palumbo V, Calandriello M, Crestani A, Zuiani C, Ficarra V (2018) Inter-reader agreement of Prostate Imaging Reporting and Data System version 2 in detecting prostate cancer on 3 Tesla multiparametric MRI: A prospective study on patients referred to radical prostatectomy. Eur Urol Suppl 17:e893.
Vaché T, Bratan F, Mège-Lechevallier F, Roche S, Rabilloud M, Rouvière O (2014) Characterization of Prostate Lesions as Benign or Malignant at Multiparametric MR Imaging: Comparison of Three Scoring Systems in Patients Treated with Radical Prostatectomy. Radiology 272:446–455.
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444.
Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J (2016) Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans Med Imaging 35:1299–1312.
Wu S, Zhong S, Liu Y (2017) Deep residual learning for image steganalysis. Multimed Tools Appl 1–17.
Le MH, Chen J, Wang L, Wang Z, Liu W, Cheng K-T (Tim), Yang X (2017) Automated diagnosis of prostate cancer in multi-parametric MRI based on multimodal convolutional neural networks. Phys Med Biol 62:6497–6514.
Krizhevsky A (2009) Learning Multiple Layers of Features from Tiny Images.
Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, Guadarrama S, Darrell T (2014) Caffe: Convolutional Architecture for Fast Feature Embedding.
Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM (2016) Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning.
He K, Zhang X, Ren S, Sun J (2015) Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification.
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:77.
R Core Team (2014) R: A language and environment for statistical computing. R Found Stat Comput Vienna, Austria 2014.
Zhao C, Gao G, Fang D, Li F, Yang X, Wang H, He Q, Wang X (2016) The efficiency of multiparametric magnetic resonance imaging (mpMRI) using PI-RADS Version 2 in the diagnosis of clinically significant prostate cancer. https://doi.org/10.1016/j.clinimag.2016.04.010
Breiman L (2001) Random forests. Mach Learn 45:5–32.
Ishioka J, Matsuoka Y, Uehara S, Yasuda Y, Kijima T, Yoshida S, Yokoyama M, Saito K, Kihara K, Numao N, Kimura T, Kudo K, Kumazawa I, Fujii Y (2018) Computer-aided diagnosis of prostate cancer on magnetic resonance imaging using a convolutional neural network algorithm. BJU Int 122:411–417.
Acknowledgements
The study was supported by Siemens Medical Solutions and by funding from the Integrated Diagnostics program of the Departments of Radiological Sciences & Pathology & Laboratory Medicine, David Geffen School of Medicine at UCLA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare no conflict of interest.
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Since the study involved purely retrospective analysis of previously acquired data, the Institutional Review Board waived the need for additional informed consent.
Rights and permissions
About this article
Cite this article
Zhong, X., Cao, R., Shakeri, S. et al. Deep transfer learning-based prostate cancer classification using 3 Tesla multi-parametric MRI. Abdom Radiol 44, 2030–2039 (2019). https://doi.org/10.1007/s00261-018-1824-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-018-1824-5