Abstract
Purpose
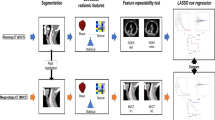
This study aimed to investigate the impact of several ComBat harmonization strategies, intra-tumoral sub-volume characterization, and automatic segmentations for progression-free survival (PFS) prediction through radiomics modeling for patients with head and neck cancer (HNC) in PET/CT images.
Methods
The HECKTOR MICCAI 2021 challenge set containing PET/CT images and clinical data of 325 oropharynx HNC patients was exploited. A total of 346 IBSI-compliant radiomic features were extracted for each patient’s primary tumor volume defined by the reference manual contours. Modeling relied on least absolute shrinkage Cox regression (Lasso-Cox) for feature selection (FS) and Cox proportional-hazards (CoxPH) models were built to predict PFS. Within this methodological framework, 8 different strategies for ComBat harmonization were compared, including before or after FS, in feature groups separately or all features directly, and with center or clustering-determined labels. Features extracted from tumor sub-volume clustering were also investigated for their prognostic additional value. Finally, 3 automatic segmentations (2 threshold-based and a 3D U-Net) were also compared. All results were evaluated with the concordance index (C-index).
Results
Radiomics features without harmonization, combined with clinical factors, led to models with C-index values of 0.69 in the testing set. The best version of ComBat harmonization, i.e., after FS, for feature groups separately and relying on clustering-determined labels, achieved a C-index of 0.71. The use of features extracted from tumor sub-volumes further improved the C-index to 0.72. Models that relied on the automatic segmentations yielded close but slightly lower prognostic performance (0.67–0.70) compared to reference contours.
Conclusion
A standard radiomics pipeline allowed for prediction of PFS in a multicenter HNC cohort. Applying a specific strategy of ComBat harmonization improved the performance. The extraction of intra-tumoral sub-volume features and automatic segmentation could contribute to the improvement and automation of prognosis modeling, respectively.






Similar content being viewed by others
Data availability
Datasets are available through the challenge website of https://www.aicrowd.com/challenges/miccai-2021-hecktor.
Code availability
Codes are available from the corresponding author on reasonable request.
References
Chow L. Head and neck cancer. N Engl J Med. 2020. https://doi.org/10.1056/NEJMra1715715.
Hatt M, Majdoub M, Vallieres M, Tixier F, Le Rest CC, Groheux D, et al. 18F-FDG PET uptake characterization through texture analysis: investigating the complementary nature of heterogeneity and functional tumor volume in a multi-cancer site patient cohort. J Nucl Med. 2015. https://doi.org/10.2967/jnumed.114.144055.
Lambin P, Leijenaar R, Deist TM, Peerlings J, de Jong E, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017. https://doi.org/10.1038/nrclinonc.2017.141.
Hatt M, Krizsan AK, Rahmim A, Bradshaw TJ, Costa PF, Forgacs A, et al. Joint EANM/SNMMI guideline on radiomics in nuclear medicine. Eur J Nucl Med Mol I. 2022. https://doi.org/10.1007/s00259-022-06001-6.
Hatt M, Cheze LRC, Antonorsi N, Tixier F, Tankyevych O, Jaouen V, et al. Radiomics in PET/CT: current status and future AI-based evolutions. Semin Nucl Med. 2021. https://doi.org/10.1053/j.semnuclmed.2020.09.002.
Hatt M, Le Rest CC, Tixier F, Badic B, Schick U, Visvikis D. Radiomics: data are also images. J Nucl Med. 2019. https://doi.org/10.2967/jnumed.118.220582.
Papadimitroulas P, Brocki L, Chung NC, Marchadour W, Vermet F, Gaubert L, et al. Artificial intelligence: deep learning in oncological radiomics and challenges of interpretability and data harmonization. Physica Med. 2021. https://doi.org/10.1016/j.ejmp.2021.03.009.
Yan J, Chu-Shern JL, Loi HY, Khor LK, Sinha AK, Quek ST, et al. Impact of image reconstruction settings on texture features in 18F-FDG PET. J Nucl Med. 2015. https://doi.org/10.2967/jnumed.115.156927.
Boellaard R, Delgado-Bolton R, Oyen WJ, Giammarile F, Tatsch K, Eschner W, et al. FDG PET/CT: EANM procedure guidelines for tumour imaging: version 2.0. Eur J Nucl Med Mol Imaging. 2015; https://doi.org/10.1007/s00259-014-2961-x.
Pfaehler E, van Sluis J, Merema BB, van Ooijen P, Berendsen RC, van Velden FH, et al. Experimental multicenter and multivendor evaluation of the performance of PET radiomic features using 3-dimensionally printed phantom inserts. J Nucl Med. 2020. https://doi.org/10.2967/jnumed.119.229724.
Llera A, Huertas I, Mir P, Beckmann CF. Quantitative intensity harmonization of dopamine transporter SPECT images using gamma mixture models. Mol Imaging Biol. 2019. https://doi.org/10.1007/s11307-018-1217-8.
Marcadent S, Hofmeister J, Preti MG, Martin SP, Van De Ville D, Montet X. Generative adversarial networks improve the reproducibility and discriminative power of radiomic features. Radiol Artif Intell. 2020. https://doi.org/10.1148/ryai.2020190035.
Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007. https://doi.org/10.1093/biostatistics/kxj037.
Orlhac F, Frouin F, Nioche C, Ayache N, Buvat I. Validation of a method to compensate multicenter effects affecting CT radiomics. Radiology. 2019. https://doi.org/10.1148/radiol.2019182023.
Orlhac F, Boughdad S, Philippe C, Stalla-Bourdillon H, Nioche C, Champion L, et al. A postreconstruction harmonization method for multicenter radiomic studies in PET. J Nucl Med. 2018. https://doi.org/10.2967/jnumed.117.199935.
Orlhac F, Eertink JJ, Cottereau AS, Zijlstra JM, Thieblemont C, Meignan M, et al. A guide to ComBat harmonization of imaging biomarkers in multicenter studies. J Nucl Med. 2022. https://doi.org/10.2967/jnumed.121.262464.
Da-ano R, Masson I, Lucia F, Doré M, Robin P, Alfieri J, et al. Performance comparison of modified ComBat for harmonization of radiomic features for multicenter studies. Sci Rep. 2020. https://doi.org/10.1038/s41598-020-66110-w.
O’Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res. 2015. https://doi.org/10.1158/1078-0432.CCR-14-0990.
Wu J, Gensheimer MF, Zhang N, Guo M, Liang R, Zhang C, et al. Tumor subregion evolution-based imaging features to assess early response and predict prognosis in oropharyngeal cancer. J Nucl Med. 2020. https://doi.org/10.2967/jnumed.119.230037.
Xu H, Lv W, Feng H, Du D, Yuan Q, Wang Q, et al. Subregional radiomics analysis of PET/CT imaging with intratumor partitioning: application to prognosis for nasopharyngeal carcinoma. Mol Imaging Biol. 2020. https://doi.org/10.1007/s11307-019-01439-x.
Vallieres M, Zwanenburg A, Badic B, Cheze LRC, Visvikis D, Hatt M. Responsible radiomics research for faster clinical translation. J Nucl Med. 2018. https://doi.org/10.2967/jnumed.117.200501.
Andrearczyk V, Oreiller V, Boughdad S, Rest CCL, Elhalawani H, Jreige M, et al. Overview of the HECKTOR challenge at MICCAI 2021: automatic head and neck tumor segmentation and outcome prediction in PET/CT images. In: 3D head and neck tumor segmentation in PET/CT challenge. Springer; 2021. pp. 1–37. https://doi.org/10.1007/978-3-030-98253-9_1.
Zwanenburg A, Vallières M, Abdalah MA, Aerts HJ, Andrearczyk V, Apte A, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020. https://doi.org/10.1148/radiol.2020191145.
Iantsen A, Visvikis D, Hatt M. Squeeze-and-excitation normalization for automated delineation of head and neck primary tumors in combined PET and CT images. In: 3D head and neck tumor segmentation in PET/CT challenge. Springer; 2020. pp. 37–43. https://doi.org/10.1007/978-3-030-67194-5_4.
Kapp AV, Tibshirani R. Are clusters found in one dataset present in another dataset? Biostatistics. 2007. https://doi.org/10.1093/biostatistics/kxj029.
Efron B, Hastie T. Computer age statistical inference. Cambridge University Press. 2016. https://doi.org/10.1017/CBO9781316576533.
Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Journal of British Surgery. 2015. https://doi.org/10.1136/bmj.g7594.
Salmanpour MR, Hajianfar G, Rezaeijo SM, Ghaemi M, Rahmim A. Advanced automatic segmentation of tumors and survival prediction in head and neck cancer. In: 3D head and neck tumor segmentation in PET/CT challenge. Springer; 2021. pp. 202–210. https://doi.org/10.1007/978-3-030-98253-9_19.
Starke S, Thalmeier D, Steinbach P, Piraud M. A hybrid radiomics approach to modeling progression-free survival in head and neck cancers. In: 3D head and neck tumor segmentation in PET/CT challenge. Springer; 2021. pp. 266–277. https://doi.org/10.1007/978-3-030-98253-9_25.
Saeed N, Majzoub RA, Sobirov I, Yaqub M. An ensemble approach for patient prognosis of head and neck tumor using multimodal data. In: 3D head and neck tumor segmentation in PET/CT challenge. Springer; 2021. pp. 278–286. https://doi.org/10.1007/978-3-030-98253-9_26.
Murugesan GK, Brunner E, McCrumb D, Kumar J, VanOss J, Moore S, et al. Head and neck primary tumor segmentation using deep neural networks and adaptive ensembling. In: 3D head and neck tumor segmentation in PET/CT challenge. Springer; 2021. pp. 224–235. https://doi.org/10.1007/978-3-030-98253-9_21.
Naser MA, Wahid KA, Mohamed AS, Abdelaal MA, He R, Dede C, et al. Progression free survival prediction for head and neck cancer using deep learning based on clinical and PET/CT imaging data. In: 3D head and neck tumor segmentation in PET/CT challenge. Springer; 2021. pp. 287–299. https://doi.org/10.1007/978-3-030-98253-9_27.
Nagpal C, Yadlowsky S, Rostamzadeh N, Heller K. Deep Cox mixtures for survival regression. In: Machine learning for healthcare conference. PMLR; 2021. pp. 674–708. https://doi.org/10.48550/arXiv.2101.06536.
Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. The annals of applied statistics. 2008. https://doi.org/10.1214/08-AOAS169.
Bogowicz M, Riesterer O, Stark LS, Studer G, Unkelbach J, Guckenberger M, et al. Comparison of PET and CT radiomics for prediction of local tumor control in head and neck squamous cell carcinoma. Acta Oncol. 2017. https://doi.org/10.1080/0284186X.2017.1346382.
Ferreira M, Lovinfosse P, Hermesse J, Decuypere M, Rousseau C, Lucia F, et al. [18F] FDG PET radiomics to predict disease-free survival in cervical cancer: a multi-scanner/center study with external validation. Eur J Nucl Med Mol I. 2021. https://doi.org/10.1007/s00259-021-05397-x.
Chatterjee A, Vallières M, Dohan A, Levesque IR, Ueno Y, Saif S, et al. Creating robust predictive radiomic models for data from independent institutions using normalization. IEEE Transactions on Radiation and Plasma Medical Sciences. 2019. https://doi.org/10.1109/TRPMS.2019.2893860.
Chen C, Grennan K, Badner J, Zhang D, Gershon E, Jin L, et al. Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PLoS ONE. 2011. https://doi.org/10.1371/journal.pone.0017238.
Masson I, Da-ano R, Lucia F, Doré M, Castelli J, Goislard De Monsabert C, et al. Statistical harmonization can improve the development of a multicenter CT-based radiomic model predictive of nonresponse to induction chemotherapy in laryngeal cancers. Med Phys. 2021; https://doi.org/10.1002/mp.14948.
Fontaine P, Andrearczyk V, Oreiller V, Abler D, Castelli J, Acosta O, et al. Cleaning radiotherapy contours for radiomics studies, is it worth it? A head and neck cancer study. Clinical and Translational Radiation Oncology. 2022. https://doi.org/10.1016/j.ctro.2022.01.003.
Sepehri S, Tankyevych O, Iantsen A, Visvikis D, Hatt M, Le Rest CC. Accurate tumor delineation vs. rough volume of interest analysis for 18F-FDG PET/CT radiomics-based prognostic modeling in non-small cell lung cancer. Front Oncol. 2021. https://doi.org/10.3389/fonc.2021.726865.
Pietras K, Östman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010. https://doi.org/10.1016/j.yexcr.2010.02.045.
Acknowledgements
We thank the organizers of the HECKTOR 2021 challenge for authorizing the use of the dataset.
Funding
This work was partly funded by (1) regions Bretagne, Pays de la Loire et Centre through the project HARMONY of the Canceropole Grand Ouest; and (2) the National Natural Science Foundation of China under grants 81871437 and 12026601, and the China Scholarship Council under grant 202108440348.
Author information
Authors and Affiliations
Contributions
Hui Xu, Nassib Abdallah, Jean-Marie Marion, Pierre Chauvet, Clovis Tauber, and Thomas Carlier searched relevant literatures and collected the data. Hui Xu, Lijun Lu, and Mathieu Hatt designed this study. Hui Xu, Nassib Abdallah, and Mathieu Hatt performed the data analysis and interpretation. Hui Xu and Mathieu Hatt drafted the primary manuscript, and all authors edited and reviewed it.
Corresponding author
Ethics declarations
Ethics approval
This is a retrospective study of a publicly available dataset. The requirement of informed consent was waived.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Oncology—Head and Neck.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, H., Abdallah, N., Marion, JM. et al. Radiomics prognostic analysis of PET/CT images in a multicenter head and neck cancer cohort: investigating ComBat strategies, sub-volume characterization, and automatic segmentation. Eur J Nucl Med Mol Imaging 50, 1720–1734 (2023). https://doi.org/10.1007/s00259-023-06118-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-023-06118-2