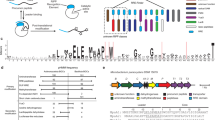
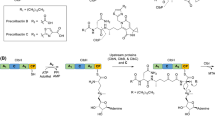
Abstract
Non-ribosomal peptide synthetases are mega-enzyme assembly lines that synthesize many clinically useful compounds. As a gatekeeper, they have an adenylation (A)-domain that controls substrate specificity and plays an important role in product structural diversity. This review summarizes the natural distribution, catalytic mechanism, substrate prediction methods, and in vitro biochemical analysis of the A-domain. Taking genome mining of polyamino acid synthetases as an example, we introduce research on mining non-ribosomal peptides based on A-domains. We discuss how non-ribosomal peptide synthetases can be engineered based on the A-domain to obtain novel non-ribosomal peptides. This work provides guidance for screening non-ribosomal peptide-producing strains, offers a method to discover and identify A-domain functions, and will accelerate the engineering and genome mining of non-ribosomal peptide synthetases.
Key points
• Introducing adenylation domain structure, substrate prediction, and biochemical analysis methods
• Advances in mining homo polyamino acids based on adenylation domain analysis
• Creating new non-ribosomal peptides by engineering adenylation domains
Graphical Abstract





Similar content being viewed by others
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Bachmann BO, Ravel J (2009) Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from dna sequence data. In: Hopwood DA (ed) Complex enzymes in microbial natural product biosynthesis, part A: overview articles and peptides. Elsevier Academic Press Inc, San Diego, pp 181–217
Bankar SB, Nimbalkar PR, Chavan PV, Singhal RS (2018) Microbial polyamino acids: an overview for commercial attention. In: Grumezescu AM, Holban AM (eds) Role of materials science in food bioengineering. Academic Press, New York, pp 381–412
Baranasic D, Zucko J, Diminic J, Gacesa R, Long PF, Cullum J, Hranueli D, Starcevic A (2014) Predicting substrate specificity of adenylation domains of nonribosomal peptide synthetases and other protein properties by latent semantic indexing. J Ind Microbiol Biotechnol 41(2):461–467. https://doi.org/10.1007/s10295-013-1322-2
Beld J, Sonnenschein EC, Vickery CR, Noel JP, Burkart MD (2014) The phosphopantetheinyl transferases: catalysis of a post-translational modification crucial for life. Nat Prod Rep 31(1):61–108. https://doi.org/10.1039/c3np70054b
Belshaw PJ, Walsh CT, Stachelhaus T (1999) Aminoacyl-CoAs as probes of condensation domain selectivity in nonribosomal peptide synthesis. Science 284(5413):486–489. https://doi.org/10.1126/science.284.5413.486
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47(W1):W81–W87. https://doi.org/10.1093/nar/gkz310
Bozhuyuk KA, Micklefield J, Wilkinson B (2019) Engineering enzymatic assembly lines to produce new antibiotics. Curr Opin Microbiol 51:88–96. https://doi.org/10.1016/j.mib.2019.10.007
Brown AS, Calcott MJ, Collins VM, Owen JG, Ackerley DF (2020) The indigoidine synthetase BpsA provides a colorimetric ATP assay that can be adapted to quantify the substrate preferences of other NRPS enzymes. Biotechnol Lett 42(12):2665–2671. https://doi.org/10.1007/s10529-020-02972-4
Calcott MJ, Owen JG, Ackerley DF (2020) Efficient rational modification of non-ribosomal peptides by adenylation domain substitution. Nat Commun 11(1):4554. https://doi.org/10.1038/s41467-020-18365-0
Chavali AK, Rhee SY (2018) Bioinformatics tools for the identification of gene clusters that biosynthesize specialized metabolites. Briefings Bioinf 19(5):1022–1034. https://doi.org/10.1093/bib/bbx020
Conti E, Stachelhaus T, Marahiel MA, Brick P (1997) Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J 16(14):4174–4183. https://doi.org/10.1093/emboj/16.14.4174
Covington BC, Xu F, Seyedsayamdost MR (2021) A natural product chemist’s guide to unlocking silent biosynthetic gene clusters. Annu Rev Biochem 90:763–788. https://doi.org/10.1146/annurev-biochem-081420-102432
D’Ambrosio HK, Derbyshire ER (2020) Investigating the role of class I adenylate-forming enzymes in natural product biosynthesis. ACS Chem Biol 15(1):17–27. https://doi.org/10.1021/acschembio.9b00865
Dekimpe S, Masschelein J (2021) Beyond peptide bond formation: the versatile role of condensation domains in natural product biosynthesis. Nat Prod Rep 38(10):1910–1937. https://doi.org/10.1039/d0np00098a
Drake EJ, Miller BR, Shi C, Tarrasch JT, Sundlov JA, Allen CL, Skiniotis G, Aldrich CC, Gulick AM (2016) Structures of two distinct conformations of holo-non-ribosomal peptide synthetases. Nature 529(7585):235–238. https://doi.org/10.1038/nature16163
Ehmann DE, Trauger JW, Stachelhaus T, Walsh CT (2000) Aminoacyl-SNACs as small-molecule substrates for the condensation domains of nonribosomal peptide synthetases. Chem Biol 7(10):765–772. https://doi.org/10.1016/s1074-5521(00)00022-3
Felnagle EA, Jackson EE, Chan YA, Podevels AM, Berti AD, McMahon MD, Thomas MG (2008) Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol Pharmaceutics 5(2):191–211. https://doi.org/10.1021/mp700137g
Horsman ME, Hari TP, Boddy CN (2016) Polyketide synthase and non-ribosomal peptide synthetase thioesterase selectivity: logic gate or a victim of fate? Nat Prod Rep 33(2):183–202. https://doi.org/10.1039/c4np00148f
Hwang S, Lee N, Cho S, Palsson B, Cho BK (2020) Repurposing modular polyketide synthases and non-ribosomal peptide synthetases for novel chemical biosynthesis. Front Mol Biosci 7:87. https://doi.org/10.3389/fmolb.2020.00087
Ishikawa F, Konno S, Uchida C, Suzuki T, Takashima K, Dohmae N, Kakeya H, Tanabe G (2022) Chemoproteomics profiling of surfactin-producing nonribosomal peptide synthetases in living bacterial cells. Cell Chem Biol 29(1):145–156. https://doi.org/10.1016/j.chembiol.2021.05.014
Ishikawa F, Tanabe G (2019) Chemical strategies for visualizing and analyzing endogenous nonribosomal peptide synthetase (nrps) megasynthetases. ChemBioChem 20(16):2032–2040. https://doi.org/10.1002/cbic.201900186
Ishikawa F, Tanabe G, Kakeya H (2019) Activity-based protein profiling of non-ribosomal peptide synthetases. In: Cravatt BF, Hsu KL, Weerapana E (eds) Activity-based protein profiling. Springer International Publishing Ag, Cham, pp 321–349
Izore T, Candace Ho YT, Kaczmarski JA, Gavriilidou A, Chow KH, Steer DL, Goode RJA, Schittenhelm RB, Tailhades J, Tosin M, Challis GL, Krenske EH, Ziemert N, Jackson CJ, Cryle MJ (2021) Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity. Nat Commun 12(1):2511. https://doi.org/10.1038/s41467-021-22623-0
Izore T, Cryle MJ (2018) The many faces and important roles of protein-protein interactions during non-ribosomal peptide synthesis. Nat Prod Rep 35(11):1120–1139. https://doi.org/10.1039/c8np00038g
Jiang XL, Radko Y, Gren T, Palazzotto E, Jorgensen TS, Cheng T, Xian M, Weber T, Lee SY (2021) Distribution of epsilon-poly-L-lysine synthetases in coryneform bacteria isolated from cheese and human skin. Appl Environ Microbiol 87(10):e01841-e1920. https://doi.org/10.1128/aem.01841-20
Kadi N, Challis GL (2009) Siderophore biosynthesis: a substrate specificity assay for nonribosomal peptide synthetase-independent siderophore synthetases involving trapping of acyl-adenylate intermediates with hydroxylamine. In: Hopwood DA (ed) Complex enzymes in microbial natural product biosynthesis, Part A: overview articles and peptides. Elsevier Academic Press Inc, San Diego, pp 431–457
Kahlert L, Lichstrahl MS, Townsend CA (2022) Colorimetric determination of adenylation domain activity in nonribosomal peptide synthetases by using chrome azurol s. Chembiochem 24(5):e202200668. https://doi.org/10.1002/cbic.202200668
Kasai S, Konno S, Ishikawa F, Kakeya H (2015) Functional profiling of adenylation domains in nonribosomal peptide synthetases by competitive activity-based protein profiling. Chem Commun 51(87):15764–15767. https://doi.org/10.1039/c5cc04953a
Katano H, Watanabe H, Takakuwa M, Maruyama C, Hamano Y (2013) Colorimetric determination of pyrophosphate anion and its application to adenylation enzyme assay. Anal Sci 29(11):1095–1098. https://doi.org/10.2116/analsci.29.1095
Kittila T, Schoppet M, Cryle MJ (2016) Online pyrophosphate assay for analyzing adenylation domains of nonribosomal peptide synthetases. ChemBioChem 17(7):576–584. https://doi.org/10.1002/cbic.201500555
Konno S, Ishikawa F, Suzuki T, Dohmae N, Burkart MD, Kakeya HJCC (2015) Active site-directed proteomic probes for adenylation domains in nonribosomal peptide synthetases. Chem Commun 51(12):2262–2265. https://doi.org/10.1039/c4cc09412c
Kranz J, Wenski SL, Dichter AA, BodeBozhüyük HBKA (2021) Influence of condensation domains on activity and specificity of adenylation domains. BioRxiv. https://doi.org/10.1101/2021.08.23.457306
Leclère V, Weber T, Jacques P, Pupin M (2016) Bioinformatics tools for the discovery of new nonribosomal peptides. In: Evans BS (ed) In: Nonribosomal peptide and polyketide biosynthesis: Methods and Protocols. Humana Press Inc., Totowa, pp 209–232
Lee SG, Lipmann F (1975) Tyrocidine synthetase system. In: Hash JH (ed) Methods in enzymology XLIII. Academic Press, New York, pp 585–602
Lee TV, Johnson RD, Arcus VL, Lott JS (2015) Prediction of the substrate for nonribosomal peptide synthetase (NRPS) adenylation domains by virtual screening. Proteins 83(11):2052–2066. https://doi.org/10.1002/prot.24922
Li YX, Zhong Z, Zhang WP, Qian PY (2018) Discovery of cationic nonribosomal peptides as Gram-negative antibiotics through global genome mining. Nat Commun 9(1):3273. https://doi.org/10.1038/s41467-018-05781-6
Liu D, Rubin GM, Dhakal D, Chen M, Ding Y (2021) Biocatalytic synthesis of peptidic natural products and related analogues. IScience 24(5):102512. https://doi.org/10.1016/j.isci.2021.102512
McQuade TJ, Shallop AD, Sheoran A, Delproposto JE, Tsodikov OV, Garneau-Tsodikova S (2009) A nonradioactive high-throughput assay for screening and characterization of adenylation domains for nonribosomal peptide combinatorial biosynthesis. Anal Biochem 386(2):244–250. https://doi.org/10.1016/j.ab.2008.12.014
Miyanaga A, Kudo F, Eguchi T (2022) Recent advances in the structural analysis of adenylation domains in natural product biosynthesis. Curr Opin Chem Biol 71:102212. https://doi.org/10.1016/j.cbpa.2022.102212
Morgan GL, Kretsch AM, Santa Maria KC, Weeks SJ, Li B (2019) Specificity of nonribosomal peptide synthetases in the biosynthesis of the pseudomonas virulence factor. Biochemistry 58(52):5249–5254. https://doi.org/10.1021/acs.biochem.9b00360
Paysan-Lafosse T, Blum M, Chuguransky S, Grego T, Pinto BL, Salazar GA, Bileschi ML, Bork P, Bridge A, Colwell L, Gough J, Haft DH, Letunic I, Marchler-Bauer A, Mi H, Natale DA, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A (2023) InterPro in 2022. Nucleic Acids Res 51(D1):D418–D427. https://doi.org/10.1093/nar/gkac993
Pham JV, Yilma MA, Feliz A, Majid MT, Maffetone N, Walker JR, Kim E, Cho HJ, Reynolds JM, Song MCJFim, (2019) A review of the microbial production of bioactive natural products and biologics. Front Microbiol 10:1404. https://doi.org/10.3389/fmicb.2019.01404
Phelan VV, Du Y, McLean JA, Bachmann BO (2009) Adenylation enzyme characterization using γ-18O4-ATP pyrophosphate exchange. Chem Biol 16(5):473–478. https://doi.org/10.1016/j.chembiol.2009.04.007
Prieto C, Garcia-Estrada C, Lorenzana D, Martin JF (2012) NRPSsp: non-ribosomal peptide synthase substrate predictor. Bioinformatics 28(3):426–427. https://doi.org/10.1093/bioinformatics/btr659
Reimer JM, Hague AS, Tarry MJ, Schmeing TM (2018) Piecing together nonribosomal peptide synthesis. Curr Opin Struc Biol 49:104–113. https://doi.org/10.1016/j.sbi.2018.01.011
Rottig M, Medema MH, Blin K, Weber T, Rausch C, Kohlbacher O (2011) NRPSpredictor2-a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res 39:W362–W367. https://doi.org/10.1093/nar/gkr323
Schmelz S, Naismith JH (2009) Adenylate-forming enzymes. Curr Opin Struct Biol 19(6):666–671. https://doi.org/10.1016/j.sbi.2009.09.004
Stachelhaus T, Mootz HD, Marahiel MA (1999) The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol 6(8):493–505. https://doi.org/10.1016/s1074-5521(99)80082-9
Stanisic A, Husken A, Kries H (2019) HAMA: a multiplexed LC-MS/MS assay for specificity profiling of adenylate-forming enzymes. Chem Sci 10(44):10395–10399. https://doi.org/10.1039/c9sc04222a
Stanisic A, Kries H (2019) Adenylation domains in nonribosomal peptide engineering. ChemBioChem 20(11):1347–1356. https://doi.org/10.1002/cbic.201800750
Throckmorton K, Vinnik V, Chowdhury R, Cook T, Chevrette MG, Maranas C, Pfleger B, Thomas MG (2019) Directed evolution reveals the functional sequence space of an adenylation domain specificity code. ACS Chem Biol 14(9):2044–2054. https://doi.org/10.1021/acschembio.9b00532
Walsh CT, Brien RVO, Khosla C (2013) Nonproteinogenic amino acid building blocks for nonribosomal peptide and hybrid polyketide scaffolds. Angew Chem Int Edit 52(28):7098–7124. https://doi.org/10.1002/anie.201208344
Weber T, Kim HU (2016) The secondary metabolite bioinformatics portal: computational tools to facilitate synthetic biology of secondary metabolite production. Synth Syst Biotechnol 1(2):69–79. https://doi.org/10.1016/j.synbio.2015.12.002
Wenski SL, Thiengmag S, Helfrich EJN (2022) Complex peptide natural products: biosynthetic principles, challenges and opportunities for pathway engineering. Synth Syst Biotechnol 7(1):631–647. https://doi.org/10.1016/j.synbio.2022.01.007
Wilson DJ, Aldrich CC (2010) A continuous kinetic assay for adenylation enzyme activity and inhibition. Anal Biochem 404(1):56–63. https://doi.org/10.1016/j.ab.2010.04.033
Winn M, Fyans JK, Zhuo Y, Micklefield J (2016) Recent advances in engineering nonribosomal peptide assembly lines. Nat Prod Rep 33(2):317–347. https://doi.org/10.1039/C5NP00099H
Xu D, Wang R, Xu Z, Xu Z, Li S, Wang M, Feng X, Xu H (2019) Discovery of a short-chain ε-poly-L-lysine and its highly efficient production via synthetase swap strategy. J Agric Food Chem 67(5):1453–1462. https://doi.org/10.1021/acs.jafc.8b06019
Xu Z, Sun Z, Li S, Xu Z, Cao C, Xu Z, Feng X, Xu H (2015) Systematic unravelling of the biosynthesis of poly (L-diaminopropionic acid) in Streptomyces albulus PD-1. Sci Rep 514(5):17400. https://doi.org/10.1038/srep17400
Xu Z, Xu Z, Feng X, Xu D, Liang J, Xu HJAm, biotechnology, (2016) Recent advances in the biotechnological production of microbial poly (ɛ-L-lysine) and understanding of its biosynthetic mechanism. Appl Microbiol Biotechnol 100(15):6619–6630. https://doi.org/10.1007/s00253-016-7677-3
Yamanaka K, Fukumoto H, Takehara M, Hamano Y, Oikawa T (2020) The stereocontrolled biosynthesis of mirror-symmetric 2,4-diaminobutyric acid homopolymers is critically governed by adenylation activations. ACS Chem Biol 15(7):1964–1973. https://doi.org/10.1021/acschembio.0c00321
Yamanaka K, Fukumoto H, Yoshimura N, Arakawa K, Kato Y, Hamano Y, Oikawa T (2022) Discovery of a polyamino acid antibiotic solely comprising L-beta-lysine by potential producer prioritization-guided genome mining. ACS Chem Biol 17(1):171–180. https://doi.org/10.1021/acschembio.1c00832
Yamanaka K, Maruyama C, Takagi H, Hamano Y (2008) Epsilon-poly-L-lysine dispersity is controlled by a highly unusual nonribosomal peptide synthetase. Nat Chem Biol 4(12):766–772. https://doi.org/10.1038/nchembio.125
Funding
This work was funded by the National Nature Science Foundation of China (22208027), the Natural Science Foundation of the Jiangsu (BK20201050), the Natural Science Research Project of Jiangsu Higher Education Institutions (20KJB530019), the China Postdoctoral Science Foundation (2021M6691557), the Agricultural Science and Technology Innovation Project of Suzhou(SNG2022059), the Plan of Gusu Leading Talents of Innovation and Entrepreneurship Science and Technology (ZXL2022003), the Science and Technology Development Project of Suzhou (SNG2021018), and the Science and Technology Plan of Changshu (CS202109).
Author information
Authors and Affiliations
Contributions
DL conceived and wrote the manuscript. ZH and LY designed and generated the figures. LT, YB, ML, and HX gave critical insights on the subjects, reviewed, and improved the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This mini review does not contain any studies with human participants or animals performed by any of the authors.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, D., Zhang, Z., Yao, L. et al. Advances in the adenylation domain: discovery of diverse non-ribosomal peptides. Appl Microbiol Biotechnol 107, 4187–4197 (2023). https://doi.org/10.1007/s00253-023-12585-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-023-12585-2