Abstract
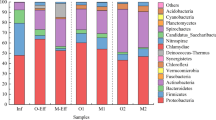
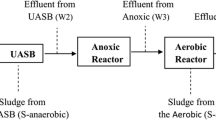
Public concerns are increasing regarding the prevalence and transmission of antibiotic resistance genes (ARGs) in wastewater treatment plants (WWTPs), especially ARG persistence and dissemination in activated sludge (AS). However, the temporal dynamics of ARGs in the AS of WWTPs over a long period of time and their transfer potential after the treatment process upgrade (e.g., total nitrogen reduction from 20 to 15 mg/L in effluent) remain poorly explored. Here, metagenomic sequencing was performed to quantify the ARGs in AS samples from two WWTPs with different treatment processes over a 2-year period. A total of 368 and 426 ARG subtypes affiliated with 20 ARG types were identified separately in the two WWTPs and the similar core ARGs were shared by all 54 samples. There were significant differences in ARG composition in different treatment processes, yet the abundance and diversity of ARGs in the AS samples demonstrated no distinct seasonal patterns. Notably, after the treatment process upgrade, the relative abundance of sulfonamide, beta-lactam, and aminoglycoside resistance genes was reduced by more than 10%, and the transfer potential of ARGs in bacterial pathogens decreased greatly, which suggested that an upgrade could limit the prevalence and transmission of ARGs. Variation partitioning analysis showed that metal resistance genes rather than bacterial community represented the significantly influential factor in shaping ARGs, and some key genera correlated with ARGs were identified through network analysis. These results will deepen our understanding of the dynamic changes in ARG profiles in AS systems and guide wastewater treatment plant upgrades.
Key points
• The potential transfer of ARGs decreased after the treatment process upgrade
• Metal resistance genes were the most influential factor in shaping ARG composition
• Co-occurrence networks displayed potential hosts of beta-lactam resistance genes





Similar content being viewed by others
Data availability
The raw sequence data of this study have been deposited at the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under the BioProject ID PRJNA798539.
References
Amachawadi RG, Scott HM, Alvarado CA, Mainini TR, Vinasco J, Drouillard JS, Nagaraja TG (2013) Occurrence of the transferable copper resistance gene tcrB among fecal enterococci of U. S. Feedlot cattle fed copper-supplemented diets. Appl Environ Microbiol 79(14):4369–4375. https://doi.org/10.1128/AEM.00503-13
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477. https://doi.org/10.1089/cmb.2012.0021
Barberán A, Bates ST, Casamayor EO, Fierer N (2012) Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6(2):343–351. https://doi.org/10.1038/ismej.2011.119
Bastian M, Heymann S, Jacomy M (2009) Gephi: an open source software for exploring and manipulating networks. Proc Third Int ICWSM Conf (ICWSM) 8:361–362
Ben W, Wang J, Cao R, Yang M, Zhang Y, Qiang Z (2017) Distribution of antibiotic resistance in the effluents of ten municipal wastewater treatment plants in China and the effect of treatment processes. Chemosphere 172:392–398. https://doi.org/10.1016/j.chemosphere.2017.01.041
Bliziotis IA, Petrosillo N, Michalopoulos A, Samonis G, Falagas ME (2011) Impact of definitive therapy with beta-lactam monotherapy or combination with an aminoglycoside or a quinolone for pseudomonas aeruginosa bacteremia. PLoS One 6(10):e26470. https://doi.org/10.1371/journal.pone.0026470
Cacace D, Fatta Kassinos D, Manaia CM, Cytryn E, Kreuzinger N, Rizzo L, Karaolia P, Schwartz T, Alexander J, Merlin C, Garelick H, Schmitt H, de Vries D, Schwermer CU, Meric S, Ozkal CB, Pons MN, Kneis D, Berendonk TU (2019) Antibiotic resistance genes in treated wastewater and in the receiving water bodies: a pan-European survey of urban settings. Water Res 162:320–330. https://doi.org/10.1016/j.watres.2019.06.039
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) Blast+: architecture and applications. BMC Bioinformatics 10(1):421. https://doi.org/10.1186/1471-2105-10-421
Cavaco LM, Hasman H, Stegger M, Andersen PS, Skov R, Fluit AC, Ito T, Aarestrup FM (2010) Cloning and occurrence of czrC, a gene conferring cadmium and zinc resistance in methicillin-resistant staphylococcus aureus cc398 isolates. Antimicrob Agents Chemother (bethesda) 54(9):3605–3608. https://doi.org/10.1128/AAC.00058-10
Chen S, Zhou Y, Chen Y, Gu J (2018) Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17):i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Chokshi A, Sifri Z, Cennimo D, Horng H (2019) Global contributors to antibiotic resistance. J Glob Infect Dis 11(1):36–42. https://doi.org/10.4103/jgid.jgid_110_18
Di Cesare A, Eckert EM, D’Urso S, Bertoni R, Gillan DC, Wattiez R, Corno G (2016) Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants. Water Res 94:208–214. https://doi.org/10.1016/j.watres.2016.02.049
Dixon P (2003) Vegan, a package of r functions for community ecology. J Veg Sci 14(6):927–930. https://doi.org/10.1111/j.1654-1103.2003.tb02228.x
Dubée V, Soroka D, Cortes M, Lefebvre A-L, Gutmann L, Hugonnet J-E, Arthur M, Mainardi J-L (2015) Impact of β-lactamase inhibition on the activity of ceftaroline against Mycobacterium tuberculosis and Mycobacterium abscessus. Antimicrob Agents Chemother (bethesda) 59(5):2938–2941. https://doi.org/10.1128/AAC.05080-14
Edgar RC (2010) Search and clustering orders of magnitude faster than blast. Bioinformatics 26(19):2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Eggen RIL, Hollender J, Joss A, Schärer M, Stamm C (2014) Reducing the discharge of micropollutants in the aquatic environment: the benefits of upgrading wastewater treatment plants. Environ Sci Technol 48(14):7683–7689. https://doi.org/10.1021/es500907n
Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N, Dantas G (2014) Bacterial phylogeny structures soil resistomes across habitats. Nature 509(7502):612–616. https://doi.org/10.1038/nature13377
French GL, Shannon KP, Simmons N (1996) Hospital outbreak of Klebsiella pneumoniae resistant to broad-spectrum cephalosporins and beta-lactam-beta-lactamase inhibitor combinations by hyperproduction of SHV-5 beta-lactamase. J Clin Microbiol 34(2):358–363. https://doi.org/10.1128/jcm.34.2.358-363.1996
Gao P, Mao D, Luo Y, Wang L, Xu B, Xu L (2012) Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res 46(7):2355–2364. https://doi.org/10.1016/j.watres.2012.02.004
Gu Z, Eils R, Schlesner M (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32(18):2847–2849. https://doi.org/10.1093/bioinformatics/btw313
Guo J, Li J, Chen H, Bond PL, Yuan Z (2017) Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res 123:468–478. https://doi.org/10.1016/j.watres.2017.07.002
Hollender J, Zimmermann SG, Koepke S, Krauss M, McArdell CS, Ort C, Singer H, von Gunten U, Siegrist H (2009) Elimination of organic micropollutants in a municipal wastewater treatment plant upgraded with a full-scale post-ozonation followed by sand filtration. Environ Sci Technol 43(20):7862–7869. https://doi.org/10.1021/es9014629
Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 11(1):119. https://doi.org/10.1186/1471-2105-11-119
Jia S, Shi P, Hu Q, Li B, Zhang T, Zhang X-X (2015) Bacterial community shift drives antibiotic resistance promotion during drinking water chlorination. Environ Sci Technol 49(20):12271–12279. https://doi.org/10.1021/acs.est.5b03521
Jiao Y-N, Zhou Z-C, Chen T, Wei Y-Y, Zheng J, Gao R-X, Chen H (2018) Biomarkers of antibiotic resistance genes during seasonal changes in wastewater treatment systems. Environ Pollut 234:79–87. https://doi.org/10.1016/j.envpol.2017.11.048
Ju F, Zhang T (2015) Bacterial assembly and temporal dynamics in activated sludge of a full-scale municipal wastewater treatment plant. ISME J 9(3):683–695. https://doi.org/10.1038/ismej.2014.162
Ju F, Beck K, Yin X, Maccagnan A, McArdell CS, Singer HP, Johnson DR, Zhang T, Bürgmann H (2019) Wastewater treatment plant resistomes are shaped by bacterial composition, genetic exchange, and upregulated expression in the effluent microbiomes. ISME J 13(2):346–360. https://doi.org/10.1038/s41396-018-0277-8
Karkman A, Johnson TA, Lyra C, Stedtfeld RD, Tamminen M, Tiedje JM, Virta M (2016) High-throughput quantification of antibiotic resistance genes from an urban wastewater treatment plant. FEMS Microbiol Ecol 92(3). https://doi.org/10.1093/femsec/fiw014
Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N (2010) Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10(9):597–602. https://doi.org/10.1016/S1473-3099(10)70143-2
Li B, Ju F, Cai L, Zhang T (2015) Profile and fate of bacterial pathogens in sewage treatment plants revealed by high-throughput metagenomic approach. Environ Sci Technol 49(17):10492–10502. https://doi.org/10.1021/acs.est.5b02345
Li B, Yang Y, Ma L, Ju F, Guo F, Tiedje JM, Zhang T (2015) Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J 9(11):2490–2502. https://doi.org/10.1038/ismej.2015.59
Li LG, Xia Y, Zhang T (2017) Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J 11(3):651–662. https://doi.org/10.1038/ismej.2016.155
Liang J, Mao G, Yin X, Ma L, Liu L, Bai Y, Zhang T, Qu J (2020) Identification and quantification of bacterial genomes carrying antibiotic resistance genes and virulence factor genes for aquatic microbiological risk assessment. Water Res 168:115160. https://doi.org/10.1016/j.watres.2019.115160
Liu Z, Klümper U, Liu Y, Yang Y, Wei Q, Lin JG, Gu JD, Li M (2019) Metagenomic and metatranscriptomic analyses reveal activity and hosts of antibiotic resistance genes in activated sludge. Environ Int 129:208–220. https://doi.org/10.1016/j.envint.2019.05.036
Lorenzo P, Adriana A, Jessica S, Carles B, Marinella F, Marta L, Luis BJ, Pierre S (2018) Antibiotic resistance in urban and hospital wastewaters and their impact on a receiving freshwater ecosystem. Chemosphere 206:70–82. https://doi.org/10.1016/j.chemosphere.2018.04.163
McKinney CW, Loftin KA, Meyer MT, Davis JG, Pruden A (2010) tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence. Environ Sci Technol 44(16):6102–6109. https://doi.org/10.1021/es9038165
Munir M, Wong K, Xagoraraki I (2011) Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res 45(2):681–693. https://doi.org/10.1016/j.watres.2010.08.033
Pal C, Bengtsson-Palme J, Rensing C, Kristiansson E, Larsson DGJ (2014) Bacmet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res 42(D1):D737–D743. https://doi.org/10.1093/nar/gkt1252
Pärnänen K, Karkman A, Hultman J, Lyra C, Bengtsson-Palme J, Larsson DGJ, Rautava S, Isolauri E, Salminen S, Kumar H, Satokari R, Virta M (2018) Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat Commun 9(1). https://doi.org/10.1038/s41467-018-06393-w
Pazda M, Kumirska J, Stepnowski P, Mulkiewicz E (2019) Antibiotic resistance genes identified in wastewater treatment plant systems – a review. Sci Total Environ 697:134023. https://doi.org/10.1016/j.scitotenv.2019.134023
Rashid SS, Liu Y-Q, Zhang C (2020) Upgrading a large and centralised municipal wastewater treatment plant with sequencing batch reactor technology for integrated nutrient removal and phosphorus recovery: environmental and economic life cycle performance. Sci Total Environ 749:141465. https://doi.org/10.1016/j.scitotenv.2020.141465
Revelle W, Revelle MW (2015) Package ‘psych.’ Compr R Arch Netw 337:338
Skurnik D, Ruimy R, Ready D, Ruppe E, Bernède-Bauduin C, Djossou F, Guillemot D, Pier GB, Andremont A (2010) Is exposure to mercury a driving force for the carriage of antibiotic resistance genes? J Med Microbiol 59(7):804–807. https://doi.org/10.1099/jmm.0.017665-0
Stepanauskas R, Glenn TC, Jagoe CH, Tuckfield RC, Lindell AH, King CJ, McArthur JV (2006) Coselection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ Microbiol 8(9):1510–1514. https://doi.org/10.1111/j.1462-2920.2006.01091.x
Su JQ, An XL, Li B, Chen QL, Gillings MR, Chen H, Zhang T, Zhu YG (2017) Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China. Microbiome 5(1):84. https://doi.org/10.1186/s40168-017-0298-y
Tong J, Tang A, Wang H, Liu X, Huang Z, Wang Z, Zhang J, Wei Y, Su Y, Zhang Y (2019) Microbial community evolution and fate of antibiotic resistance genes along six different full-scale municipal wastewater treatment processes. Bioresour Technol 272:489–500. https://doi.org/10.1016/j.biortech.2018.10.079
Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N (2015) Metaphlan2 for enhanced metagenomic taxonomic profiling. Nat Methods 12(10):902–903. https://doi.org/10.1038/nmeth.3589
Wang Y, Lu J, Engelstädter J, Zhang S, Ding P, Mao L, Yuan Z, Bond PL, Guo J (2020) Non-antibiotic pharmaceuticals enhance the transmission of exogenous antibiotic resistance genes through bacterial transformation. ISME J 14(8):2179–2196. https://doi.org/10.1038/s41396-020-0679-2
Wood DE, Salzberg SL (2014) Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15(3):R46. https://doi.org/10.1186/gb-2014-15-3-r46
Xu S, Lu W, Qasim MZ (2020) High-throughput characterization of the expressed antibiotic resistance genes in sewage sludge with transcriptional analysis. Ecotoxicol Environ Saf 205:111377. https://doi.org/10.1016/j.ecoenv.2020.111377
Yang Y, Li B, Ju F, Zhang T (2013) Exploring variation of antibiotic resistance genes in activated sludge over a four-year period through a metagenomic approach. Environ Sci Technol 47(18):10197–10205. https://doi.org/10.1021/es4017365
Yang Y, Song W, Lin H, Wang W, Du L, Xing W (2018) Antibiotics and antibiotic resistance genes in global lakes: a review and meta-analysis. Environ Int 116:60–73. https://doi.org/10.1016/j.envint.2018.04.011
Yin X, Jiang XT, Chai B, Li L, Yang Y, Cole JR, Tiedje JM, Zhang T (2018) ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics 34(13):2263–2270. https://doi.org/10.1093/bioinformatics/bty053
Yin X, Deng Y, Ma L, Wang Y, Chan LYL, Zhang T (2019) Exploration of the antibiotic resistome in a wastewater treatment plant by a nine-year longitudinal metagenomic study. Environ Int 133:105270. https://doi.org/10.1016/j.envint.2019.105270
Yuan L, Li ZH, Zhang MQ, Shao W, Fan YY, Sheng GP (2019) Mercury/silver resistance genes and their association with antibiotic resistance genes and microbial community in a municipal wastewater treatment plant. Sci Total Environ 657:1014–1022. https://doi.org/10.1016/j.scitotenv.2018.12.088
Zhang T, Shao M-F, Ye L (2012) 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J 6(6):1137–1147. https://doi.org/10.1038/ismej.2011.188
Zhang H, He H, Chen S, Huang T, Lu K, Zhang Z, Wang R, Zhang X, Li H (2019) Abundance of antibiotic resistance genes and their association with bacterial communities in activated sludge of wastewater treatment plants: geographical distribution and network analysis. J Environ Sci (china) 82:24–38. https://doi.org/10.1016/j.jes.2019.02.023
Zhang N, Juneau P, Huang R, He Z, Sun B, Zhou J, Liang Y (2021) Coexistence between antibiotic resistance genes and metal resistance genes in manure-fertilized soils. Geoderma 382:114760. https://doi.org/10.1016/j.geoderma.2020.114760
Zhao R, Feng J, Liu J, Fu W, Li X, Li B (2019) Deciphering of microbial community and antibiotic resistance genes in activated sludge reactors under high selective pressure of different antibiotics. Water Res 151:388–402. https://doi.org/10.1016/j.watres.2018.12.034
Zhou S, Zhu Y, Yan Y, Wang W, Wang Y (2019) Deciphering extracellular antibiotic resistance genes (eARGs) in activated sludge by metagenome. Water Res 161:610–620. https://doi.org/10.1016/j.watres.2019.06.048
Zhu G, Wang X, Yang T, Su J, Qin Y, Wang S, Gillings M, Wang C, Ju F, Lan B, Liu C, Li H, Long X-E, Wang X, Jetten MSM, Wang Z, Zhu Y-G (2021) Air pollution could drive global dissemination of antibiotic resistance genes. ISME J 15(1):270–281. https://doi.org/10.1038/s41396-020-00780-2
Zieliński W, Korzeniewska E, Harnisz M, Drzymała J, Felis E, Bajkacz S (2021) Wastewater treatment plants as a reservoir of integrase and antibiotic resistance genes – an epidemiological threat to workers and environment. Environ Int 156:106641. https://doi.org/10.1016/j.envint.2021.106641
Funding
The authors gratefully acknowledge the support of the National Natural Science Foundation of China (41907214), the Natural Science Foundation of Guangdong Province (2022A1515011961), the Stable Support Program of Colleges and Universities in Shenzhen (20200813153536001), and the Natural Science Foundation of Shenzhen University (860–000002110245).
Author information
Authors and Affiliations
Contributions
LT: conceived of study, wrote the paper, performed research, analyzed data, contributed new methods or models; QL: performed research, analyzed data; XC: performed research, analyzed data; YW: performed research; YW: analyzed data; YM: designed study, wrote the paper, funding acquisition.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors. This study complied with all relevant ethical standards.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tian, L., Li, Q., Cai, X. et al. Dynamic distribution and potential transmission of antibiotic resistance genes in activated sludge. Appl Microbiol Biotechnol 106, 6785–6797 (2022). https://doi.org/10.1007/s00253-022-12162-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-12162-z