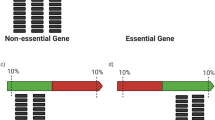
Abstract
Essential genes are crucial for bacterial viability and represent attractive targets for novel anti-pathogen drug discovery. However, essential genes determined by the transposon insertion sequencing (Tn-seq) approach often contain many false positives. We hypothesized that some of those false positives are genes that are actually deleted from the genome, so they do not present any transposon insertion in the course of Tn-seq analysis. Based on this assumption, we performed a large-scale whole-genome sequencing analysis for the bacterium of interest. Our analysis revealed that some “essential genes” are indeed removed from the analyzed bacterial genomes. Since these genes were kicked out by bacteria, they should not be defined as essential. Our work showed that gene deletion is one of the false positive sources of essentiality determination, which is apparently underestimated in previous studies. We suggest subtracting the genome backgrounds before the evaluation of Tn-seq, and created a list of false positive gene essentiality as a reference for the downstream application.
Key points
• Discovery of false positives of essential genes defined previously through the analyses of a large scale of whole-genome sequencing data
• These false positives are the results of gene deletions in the studied genomes
• Sequencing the target genome before Tn-seq analysis is of importance while some studies neglected it



Similar content being viewed by others
Data availability
SRA data files analyzed in this study are listed in Supplementary Table S1.
Code availability
Used software was listed in the section of “Materials and methods.” Codes can be found at GitHub repository (https://github.com/daiweijun/Whole-genome-sequencing-analysis.git).
References
Andrews S (2010) FastQC: A quality control tool for high throughput sequence data. Bioinformatics, Babraham Institute, Cambridge, UK
Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko K, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol 2(0008):570
Cain AK, Barquist L, Goodman AL, Paulsen IT, Parkhill J, van Opijnen T (2020) A decade of advances in transposon-insertion sequencing. Nat Rev Genet:1-15
Cameron DE, Urbach JM, Mekalanos JJ (2008) A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. PNAS 105(25):8736–8741
Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, Coller JA, Fero MJ, McAdams HH, Shapiro L (2011) The essential genome of a bacterium. Mol Syst Biol 7:528. https://doi.org/10.1038/msb.2011.58
DeJesus MA, Ioerger TR (2013) A hidden Markov model for identifying essential and growth-defect regions in bacterial genomes from transposon insertion sequencing data. BMC Bioinformatics 14(1):303
DeJesus MA, Ioerger TR (2016) Normalization of transposon-mutant library sequencing datasets to improve identification of conditionally essential genes. J Bioinf Comput Biol 14(03):1642004
DeJesus MA, Zhang YJ, Sassetti CM, Rubin EJ, Sacchettini JC, Ioerger TR (2013) Bayesian analysis of gene essentiality based on sequencing of transposon insertion libraries. Bioinformatics 29(6):695–703. https://doi.org/10.1093/bioinformatics/btt043
Dotsch A, Pommerenke C, Bredenbruch F, Geffers R, Haussler S (2009) Evaluation of a microarray-hybridization based method applicable for discovery of single nucleotide polymorphisms (SNPs) in the Pseudomonas aeruginosa genome. BMC Genomics 10:29. https://doi.org/10.1186/1471-2164-10-29
Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ (2009) Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. PNAS 106(38):16422–16427
Hutchison CA, Merryman C, Sun L, Assad-Garcia N, Richter RA, Smith HO, Glass JI (2019) Polar effects of transposon insertion into a minimal bacterial genome. J Bacteriol 201(19):e00185-e219
Judson N, Mekalanos JJ (2000) Transposon-based approaches to identify essential bacterial genes. Trends Microbiol 8(11):521–526
Juhas M, Eberl L, Church GM (2012) Essential genes as antimicrobial targets and cornerstones of synthetic biology. Trends Biotechnol 30(11):601–607
Juhas M, Eberl L, Glass JI (2011) Essence of life: essential genes of minimal genomes. Trends Cell Biol 21(10):562–568
Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, Blattner FR (2004) Systematic mutagenesis of the Escherichia coli genome. J Bacteriol 186(15):4921–4930
Klockgether J, Munder A, Neugebauer J, Davenport CF, Stanke F, Larbig KD, Heeb S, Schock U, Pohl TM, Wiehlmann L, Tummler B (2010) Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J Bacteriol 192(4):1113–1121. https://doi.org/10.1128/jb.01515-09
Koskiniemi S, Sun S, Berg OG, Andersson DI (2012) Selection-driven gene loss in bacteria. PLoS Genet 8(6):e1002787
Kuo C-H, Ochman H (2009) Deletional bias across the three domains of life. Genome Biol Evol 1:145–152
Lee SA, Gallagher LA, Thongdee M, Staudinger BJ, Lippman S, Singh PK, Manoil C (2015) General and condition-specific essential functions of Pseudomonas aeruginosa. PNAS 112(16):5189–5194
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079
Lobkovsky AE, Wolf YI, Koonin EV (2013) Gene frequency distributions reject a neutral model of genome evolution. Genome Biol Evol 5(1):233–242
Luo H, Gao F, Lin Y (2015) Evolutionary conservation analysis between the essential and nonessential genes in bacterial genomes. Sci Rep 5(1):1–8
Luo H, Lin Y, Gao F, Zhang C-T, Zhang R (2014) DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res 42(D1):D574–D580
Mira A, Ochman H, Moran NA (2001) Deletional bias and the evolution of bacterial genomes. Trends Genet 17(10):589–596. https://doi.org/10.1016/s0168-9525(01)02447-7
Murima P, McKinney JD, Pethe K (2014) Targeting bacterial central metabolism for drug development. Chem Biol 21(11):1423–1432
Pechter KB, Gallagher L, Pyles H, Manoil CS, Harwood CS (2016) Essential genome of the metabolically versatile alphaproteobacterium Rhodopseudomonas palustris. J Bacteriol 198(5):867–76. https://doi.org/10.1128/JB.00771-15
Puigbò P, Lobkovsky AE, Kristensen DM, Wolf YI, Koonin EV (2014) Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Biol 12(1):66
Rocha EP (2004) Order and disorder in bacterial genomes. Curr Opin Microbiol 7(5):519–527. https://doi.org/10.1016/j.mib.2004.08.006
Sela I, Wolf YI, Koonin EV (2016) Theory of prokaryotic genome evolution. PNAS 113(41):11399–11407
Van Opijnen T, Bodi KL, Camilli A (2009) Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6(10):767
Van Opijnen T, Camilli A (2013) Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 11(7):435–442
Zhang YJ, Loerger TR, Huttenhower C, Long JE, Sassetti CM, Sacchettini JC, Rubin EJ (2012) Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Path 8(9)
Zomer A, Burghout P, Bootsma HJ, Hermans PW, van Hijum SA (2012) ESSENTIALS: software for rapid analysis of high throughput transposon insertion sequencing data. PloS one 7(8)
Acknowledgements
Claudine Baraquet (Université de Toulon, France) is acknowledged for helpful discussing the determination of gene deletions basing on read counts. We thank E. Peter Greenberg (University of Washington, USA) for kindly donating the P. aeruginosa PAO1-UW strain. Our lab members are highly appreciable for kind supports in many aspects of this work.
Funding
This work was founded by the National Natural Science Foundation of China (31771341) and by Guangdong Province Science and Technology Innovation Strategy Special Fund (grant no: 2018B020206001).
Author information
Authors and Affiliations
Contributions
Y.L. and W.D. designed and wrote the manuscript. B.J. conducted the molecular experiments. Y.L. and W.D. performed the bioinformatic analyses. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
All authors have read and approved the final manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Y., Jiang, B. & Dai, W. A large-scale whole-genome sequencing analysis reveals false positives of bacterial essential genes. Appl Microbiol Biotechnol 106, 341–347 (2022). https://doi.org/10.1007/s00253-021-11702-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11702-3