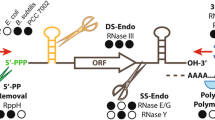
Abstract
Cyanobacteria are an ideal host for biofuel production. Although efforts have been made to genetically engineer cyanobacteria for efficient production of biofuels and other important chemicals, the tools that can be applied to cyanobacteria are still limited. A new gene regulation tool, riboregulator, has been examined for application in cyanobacteria. A riboregulator is a nature-inspired RNA tool, which is composed of two artificially designed RNA fragments. Owing to its high specificity and efficacy, it is suitable for metabolic engineering in cyanobacteria, and several studies have been done to optimize and improve the function of the riboregulator. In this review, we focus on the recent improvements made to riboregulators and compare them with other RNA-mediated gene regulation tools developed in cyanobacteria to investigate future applications of riboregulators.


Similar content being viewed by others
References
Abe K, Sakai Y, Nakashima S, Araki M, Yoshida W, Sode K, Ikebukuro K (2014) Design of riboregulators for control of cyanobacterial (Synechocystis) protein expression. Biotechnol Lett 36:287–294
Aït-Bara S, Carpousis AJ (2015) RNA degradosomes in bacteria and chloroplasts: classification, distribution and evolution of RNase E homologs. Mol Microbiol 97:1021–1135
Anders C, Niewoehner O, Duerst A, Jinek M (2014) Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513(7519):569–573
Ben Jehuda R, Shemer Y, Binah O (2018) Genome editing in induced pluripotent stem cells using CRISPR/Cas9. Stem Cell Rev 14:323–336. https://doi.org/10.1007/s12015-018-9811-3
Bøggild A, Overgaard M, Valentin-Hansen P, Brodersen DE (2009) Cyanobacteria contain a structural homologue of the Hfq protein with altered RNA-binding properties. FEBS J 276:3904–3915
Brennan RG, Link TM (2007) Hfq structure, function and ligand binding. Curr Opin Microbiol 10:125–133
Camsund D, Lindblad P (2014) Engineered transcriptional systems for cyanobacterial biotechnology. Front Bioeng Biotechnol 2:40
Chisti Y (2007) Biodiesel from microalgae. Biotechnol Adv 3:294–306
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Connor MR, Atsumi S (2010) Synthetic biology guides biofuel production. Biomed Biotechnol 2010:1–9. https://doi.org/10.1155/2010/541698
Dienst D, Dühring U, Mollenkopf HJ, Vogel J, Golecki J, Hess WR, Wilde A (2008) The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of Synechocystis sp. PCC 6803. Microbiology 154:3134–3143
Fischer S, Maier LK, Stoll B, Brendel J, Fischer E, Pfeiffer F, Dyall-Smith M, Marchfelder A (2012) An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J Biol Chem 287:33351–33363
Folichon M, Arluison V, Pellegrini O, Huntzinger E, Régnier P, Hajnsdorf E (2003) The poly(a) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res 31:7302–7310
Gordon GC, Korosh TC, Cameron JC, Markley AL, Begemann MB, Pfleger BF (2016) CRISPR interference as a titratable, trans-acting regulatory tool for metabolic engineering in the cyanobacterium Synechococcus sp. strain PCC 7002. Metab Eng 38:170–179
Heidorn T, Camsund D, HuangHH LP, Oliveira P, Stensjö K, Lindblad P (2011) Synthetic biology in cyanobacteria engineering and analyzing novel functions. Methods Enzymol 497:539–579
Hess WR, Berghoff BA, Wilde A, Steglich C, Klug G (2014) Riboregulators and the role of Hfq in photosynthetic bacteria. RNA Biol 11:413–426
Hjalt TA, Wagner EG (1995) Bulged-out nucleotides protect an antisense RNA from RNase III cleavage. Nucleic Acids Res 23:571–579
Huang HH, Camsund D, Lindblad P, Heidorn T (2010) Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res 38:2577–2593
Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ (2004) Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol 22:841–847
Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S (2015) Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol 81:2506–2514
Kaczmarzyk D, Cengic I, Yao L, Hudson EP (2018) Diversion of the long-chain acyl-ACP pool in Synechocystis to fatty alcohols through CRISPRi repression of the essential phosphate acyltransferase PlsX. Metab Eng 45:59–66
Kaniya Y, Kizawa A, Miyagi A, Kawai-Yamada M, Uchimiya H, Kaneko Y, Nishiyama Y, Hihara Y (2013) Deletion of the transcriptional regulator cyAbrB2 deregulates primary carbon metabolism in Synechocystis sp. PCC 6803. Plant Physiol 162:1153–1163
Keefer AB, Asare EK, Pomerantsev AP, Moayeri M, Martens C, Porcella SF, Gottesman S, Leppla SH, Vrentas CE (2017) In vivo characterization of an Hfq protein encoded by the Bacillus anthracis virulence plasmid pXO1. BMC Microbiol 17:63
Khan MI, Shin JH, Kim JD (2018) The promising future of microalgae: current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb Cell Factories 17:36. https://doi.org/10.1186/s12934-018-0879-x
Krishnamurthy M, Hennelly SP, Dale T, Starkenburg SR, Martí-Arbona R, Fox DT, Twary SN, Sanbonmatsu KY, Unkefer CJ (2015) Tunable riboregulator switches for post-transcriptional control of gene expression. ACS Synth Biol 4:1326–1334
Lau NS, Matsui M, Abdullah AA (2015) Cyanobacteria: photoautotrophic microbial factories for the sustainable synthesis of industrial products. Biomed Res Int 2015:754934–754939. https://doi.org/10.1155/2015/754934
Lease RA, Woodson SA (2004) Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J Mol Biol 344:1211–1223
Li Y, Lin Z, Huang C, Zhang Y, Wang Z, Tang YJ, Chen T, Zhao X (2015) Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab Eng 31:13–21
Li H, Shen CR, Huang CH, Sung LY, Wu MY, Hu YC (2016) CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production. Metab Eng 38:293–302
Link TM, Valentin-Hansen P, Brennan RG (2009) Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci U S A 106:19292–19297
López-Maury L, García-Domínguez M, Florencio FJ, Reyes JC (2002) A two-component signal transduction system involved in nickel sensing in the cyanobacterium Synechocystis sp. PCC 6803. Mol Microbiol 43:247–256
Lopez-Sanchez MJ, Sauvage E, Da Cunha V, Clermont D, Ratsima Hariniaina E, Gonzalez-Zorn B, Poyart C, Rosinski-Chupin I, Glaser P (2012) The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol Microbiol 85:1057–1071
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norvile JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
Man S, Cheng R, Miao C, Gong Q, Gu Y, Lu X, Han F, Yu W (2011) Artificial trans-encoded small non-coding RNAs specifically silence the selected gene expression in bacteria. Nucleic Acids Res 39:50. https://doi.org/10.1093/nar/gkr034
Massé E, Escorcia FE, Gottesman S (2003) Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17:2374–2383
Mazard S, Penesyan A, Ostrowski M, Paulsen IT, Egan S (2016) Tiny microbes with a big impact: the role of cyanobacteria and their metabolites in shaping our future. Mar Drugs 14:5. https://doi.org/10.3390/md14050097
Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Bläsi U (2003) Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9:1308–1314
Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, Brennan RG, Valentin-Hansen P (2002) Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell 9:23–30
Na D, Yoo SM, Chung H, Park H, Park JH, Lee SY (2013) Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol 31:170–174
Nakahira Y, Ogawa A, Asano H, Oyama T, Tozawa Y (2013) Theophylline-dependent riboswitch as a novel genetic tool for strict regulation of protein expression in cyanobacterium Synechococcus elongatus PCC 7942. Plant Cell Physiol 54:1724–1735
O’Geen H, Yu AS, Segal DJ (2015) How specific is CRISPR/Cas9 really? Curr Opin Chem Biol 29:72–78
Oh YK, Hwang KR, Kim C, Kim JR, Lee JS (2018) Recent developments and key barriers to advanced biofuels: a short review. Bioresour Technol 257:320–333
Pellagatti A, Dolatshad H, Valletta S, Boultwood J (2015) Application of CRISPR/Cas9 genome editing to the study and treatment of disease. Arch Toxicol 89:1023–1034
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183
Quintana N, Van der Kooy F, Van de Rhee MD, Voshol GP, Verpoorte R (2011) Renewable energy from cyanobacteria: energy production optimization by metabolic pathway engineering. Appl Microbiol Biotechnol 91:471–490
Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ Jr, Hallett JP, Leak DJ, Liotta CL, Mielenz JR, Murphy R, Templer R, Tschaplinski T (2006) The path forward for biofuels and biomaterials. Science 311:484–489
Sakai Y, Abe K, Nakashima S, Yoshida W, Ferri S, Sode K, Ikebukuro K (2014) Improving the gene-regulation ability of small RNAs by scaffold engineering in Escherichia coli. ACS Synth Biol 3:152–162
Sakai Y, Abe K, Nakashima S, Ellinger JJ, Ferri S, Sode K, Ikebukuro K (2015) Scaffold-fused riboregulators for enhanced gene activation in Synechocystis sp. PCC 6803. Microbiologyopen 4:533–540
Sakamoto I, Abe K, Kawai S, Tsukakoshi K, Sakai Y, Sode K, Ikebukuro K (2018) Improving the induction fold of riboregulators for cyanobacteria. RNA Biol 5:1–6
Sharma V, Yamamura A, Yokobayashi Y (2012) Engineering artificial small RNAs for conditional gene silencing in Escherichia coli. ACS Synth Biol 1:6–13
Soper TJ, Woodson SA (2008) The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 14:1907–1917
Soper TJ, Mandin P, Majdalani N, Gottesman S, Woodson SA (2010) Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci U S A 107:9602–9607
Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA (2014) DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507(7490):62–67
Sun X, Zhulin I, Wartell RM (2002) Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res 30:3662–3671
Tasan I, Zhao H (2017) Targeting specificity of the CRISPR/Cas9 system. ACS Synth Biol 6:1609–1613
Taton A, Ma AT, Ota M, Golden SS, Golden JW (2017) NOT gate genetic circuits to control gene expression in cyanobacteria. ACS Synth Biol 6:2175–2182
Ueno K, Sakai Y, Shono C, Sakamoto I, Tsukakoshi K, Hihara Y, Sode K, Ikebukuro K (2017) Applying a riboregulator as a new chromosomal gene regulation tool for higher glycogen production in Synechocystis sp. PCC 6803. Appl Microbiol Biotechnol 101:8465–8474
Wang B, Wang J, Zhang W, Meldrum DR (2012) Application of synthetic biology in cyanobacteria and algae. Front Microbiol 3:344. https://doi.org/10.3389/fmicb.2012.00344
Wang J, Wang H, Yang L, Lv L, Zhang Z, Ren B, Dong L, Li N (2018) A novel riboregulator switch system of gene expression for enhanced microbial production of succinic acid. J Ind Microbiol Biotechnol 45:253–269
Wu X, Kriz AJ, Sharp PA (2014) Target specificity of the CRISPR-Cas9 system. Quant Biol 2:59–70
Yamauchi Y, Kaniya Y, Kaneko Y, Hihara Y (2011) Physiological roles of the cyAbrB transcriptional regulator pair Sll0822 and Sll0359 in Synechocystis sp. strain PCC 6803. J Bacteriol 193:3702–3709
Yao L, Cengic I, Anfelt J, Hudson EP (2016) Multiple gene repression in cyanobacteria using CRISPRi. ACS Synth Biol 5:207–212
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Funding
This work was supported financially by the Core Research of Evolutional Science & Technology program (CREST) from the Japan Science and Technology Agency (JST).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All experiments were performed according to applicable national and institutional guidelines for the use of microorganisms.
Consent of publication
All authors approved the final version of this manuscript.
Rights and permissions
About this article
Cite this article
Ueno, K., Tsukakoshi, K. & Ikebukuro, K. Riboregulator elements as tools to engineer gene expression in cyanobacteria. Appl Microbiol Biotechnol 102, 7717–7723 (2018). https://doi.org/10.1007/s00253-018-9221-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9221-0