Abstract
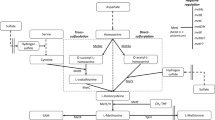
There are two alternative pathways leading to methionine synthesis in microorganisms: The transsulfuration pathway involves cystathionine as the intermediate and utilizes cysteine as the sulfur source, but the direct sulfhydrylation pathway bypasses cystathionine and uses inorganic sulfur instead. While most microorganisms synthesize methionine via either one of these pathways, Corynebacterium glutamicum utilizes both pathways, which appear to be fully functional. In C. glutamicum, each pathway is catalyzed by independent enzymes and is tightly regulated by methionine. Although the physiological significance of parallel pathways remains to be elucidated, their presence suggests metabolic flexibility and efficient adaptation of the organism to its environment.



Similar content being viewed by others
References
Alaminos M, Ramos JL (2001) The methionine biosynthetic pathway from homoserine in Pseudomonas putida involves the metW, metX, metZ, metH and meE gene products. Arch Microbiol 176:151–154
Andersen GL, Beattie GA, Lindow SE (1998) Molecular characterization and sequence of a methionine biosynthetic locus from Pseudomonas syringae. J Bacteriol 180:4497–4507
Belfaiza J, Martel A, Margarita D, Saint Girons I (1998) Direct sulfhydrylation for methionine biosynthesis in Leptospira meyeri. J Bacteriol 180:250–255
Biran D, Brot N, Weissbach H, Ron EZ (1995) Heat shock-dependent transcriptional activation of the metA gene of Escherichia coli. J Bacteriol 177:1374–1379
Biran D, Gur E, Gollan L, Ron EZ (2000) Control of methionine biosynthesis in Escherichia coli by proteolysis. Mol Microbiol 37:1436–1443
Born TL, Blanchard JS (1999) Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia coli metA-encoded homoserine transsuccinylase. Biochemistry 38:14416–14423
Born TL, Franklin M, Blanchard JS (2000) Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Haemophilus influenzae met2-encoded homoserine transacetylase. Biochemistry 39:8556–8564
Bourhy P, Martel A, Margarita D, Saint Girons I, Belfaiza J (1997) Homoserine O-acetyltransferase, involved in the Leptospira meyeri methionine biosynthetic pathway, is not feedback inhibited. J Bacteriol 179:4396–4398
Bright SWJ, Lea PJ, Miflin BJ (1979) The regulation of methionine biosynthesis and metabolism in plants and bacteria. Ciba Found Symp 72:101–117
Brzywczy J, Yamagata S, Paszewski A (1993) Comparative studies on O-acetylhomoserine sulfhydrylase: physiological role and characterization of the Aspergillus nidulans enzyme. Acta Biochim Pol 40:421–428
Brzywczy J, Sienko M, Kucharska A, Paszewski A (2002) Sulphur amino acid synthesis in Schizosaccharomyces pombe represents a specific variant of sulphur metabolism in fungi. Yeast 19:29–35
Cremer J, Treptow C, Eggeling L, Sahm H. (1988) Regulation of enzymes of lysine biosynthesis in Corynebacterium glutamicum. J Gen Microbiol 134:3221–3229
Datko AH, Giovanelli J, Mudd SH (1974) Homocysteine biosynthesis in green plants. O-Phosphorylhomoserine as the physiological substrate for cystathionine γ-synthase. J Biol Chem 249:1139–1155
Eggeling L, Sahm H (1999) l-Glutamate and l-lysine: traditional products with impetuous developments. Appl Microbiol Biotechnol 52:146–153
Eggeling L, Morbach S, Sahm H (1997) The fruits of molecular physiology: engineering the L-isoleucine biosynthesis pathway in Corynebacterium glutamicum. J Biotechnol 56:167–182
Flavin M (1971) Trans-sulfuration reactions: introduction. Methods Enzymol 17B:416–417
Flavin M, Slaughter C (1967) Enzymatic synthesis of homocysteine or methionine directly from O-succinyl-homoserine. Biochim Biophys Acta 132:400–405
Foglino M, Borne F, Bally M, Ball G, Patte JC (1995) A direct sulfhydylation pathway is used for methionine biosynthesis in Pseudomonas aeruginosa. Microbiology 141:431–439
Follettie MT, Shin HK, Sinskey AJ (1988) Organization and regulation of the Corynebacterium glutamicum hom-thrB and thrC loci. Mol Microbiol 2:53–62
Giovanelli J, Mudd SH, Datko AH (1978) Homocysteine biosynthesis in green plants. Physiological importance of the transsulfuration pathway in Chlorella sorokiniana growing under steady state conditions with limiting sulfate. J Biol Chem 253:5665–5677
Greene RC (1996) Biosynthesis of methionine. In: Escherichia coli and Salmonella: cellular and molecular biology, 2nd edition, pp. 542–560. Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger JE, eds. ASM Press, Washington DC
Großmann K, Herbster K, Mack M (2000) Rapid cloning of metK encoding methionine adenosyltransferase from Corynebacterium glutamicum by screening a genomic library on a high density colony-array. FEMS Microbiol Lett 193:99–103
Hwang B-J, Kim Y, Kim H-B, Hwang H-J, Kim J-H, Lee H-S (1999) Analysis of Corynebacterium glutamicum methionine biosynthetic pathway: isolation and analysis of metB encoding cystathionine γ-synthase. Mol Cells 9:300–308
Hwang B-J, Yeom H-J, Kim Y, Lee H-S (2002) Corynebacterium glutamicum utilizes both transsulfuration and direct sulfhydrylation pathways for methionine biosynthesis. J Bacteriol 184:1277–1286
Jetten MSM, Sinskey AJ (1995) Recent advances in the physiology and genetics of amino acid-producing bacteria. Crit Rev Biotechnol 15:73–103
Kaplan MM, Flavin M (1966) Cystathionine γ-synthetase of Salmonella. J Biol Chem 241:4463–4471
Kase H, Nakayama K (1974a) Production of O-acetyl-l-homoserine by methionine analog-resistant mutants and regulation of homoserine-O-transacetylase in Corynebacterium glutamicum. Agric Biol Chem 38:2021–2030
Kase H, Nakayama K (1974b) The regulation of l-methionine synthesis and the properties of cystathionine γ-synthase and β-cystathionase in Corynebacterium glutamicum. Agric Biol Chem 38:2235–2242
Kase H, Nakayama K (1975a) Isolation and characterization of S-adenosylmethionine-requiring mutants and role of S-adenosylmethionine in the regulation of methionine biosynthesis in Corynebacterium glutamicum. Agric Biol Chem 39:161–168
Kase H, Nakayama K (1975b) l-Methionine production by methionine analog-resistant mutants of Corynebacterium glutamicum. Agric Biol Chem 39:153–160
Kerr DS, Flavin M (1970) The regulation of methionine synthesis and the nature of cystathionine γ-synthase in Neurospora. J Biol Chem 245:1842–1855
Kertesz MA (1999) Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol Rev 24:135–175
Kim J-W, Kim H-J, Kim Y, Lee M-S, Lee H-S (2001) Properties of the Corynebacterium glutamicum metC gene encoding cystathionine β-lyase. Mol Cells 11:220–225
Kredich NM (1996) Biosynthesis of cysteine. In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger JE (eds) Escherichia coli and Salmonella: cellular and molecular biology, 2nd edn. ASM Press, Washington DC, pp. 514–527
Malumbres M, Martín JF (1996) Molecular control mechanisms of lysine and threonine biosynthesis in amino acid-producing corynebacteria: redirecting carbon flow. FEMS Microbiol Lett 143:103–114
Mares R, Urbanowski ML, Stauffer GV (1992) Regulation of the Salmonella typhimurium metA gene by the MetR protein and homocysteine. J Bacteriol 174:390–397
Martens J-H, Barg H, Warren MJ, Jahn D (2002) Microbial production of vitamin B12. Appl Microbiol Biotechnol 58:275–285
Marzluf GA (1994) Genetics and molecular genetics of sulfur assimilation in the fungi. Adv Genet 31:187–206
Marzluf GA (1997) Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu Rev Microbiol 51:73–96
Miyajima R, Shiio I (1973) Regulation of aspartate family amino acid biosynthesis in Brevibacterium flavum. VII. Properties of homoserine O-transacetylase. J Biochem 73:1061–1068
Mondal S, Chatterjee SP (1994) Enhancement of methionine production by methionine analogue ethionine resistant mutants of Brevibacterium heali. Acta Biotechnol 14:199–204
Nakamori S, Kobayashi S, Nishimura T, Takagi H (1999) Mechanism of l-methionine overproduction by Escherichia coli: the replacement of Ser-54 by Asn in the MetJ protein causes the derepression of L-methionine biosynthetic enzymes. Appl Microbiol Biotechnol 52:179–185
Neidhardt FC, Ingraham JL, Schaechter M (1990) Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Sunderland, Massachusetts, USA
O′Toole GA, Rondon MR, Trzebiatowski JR, Suh S-J, Escalante-Semerena JC (1996) Biosynthesis and utilization of adenosyl-cobalamin (coenzyme B12). In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger JE (eds) Escherichia coli and Salmonella: cellular and molecular biology, 2nd edn. ASM Press, Washington DC, pp. 542–560
Ozaki H, Shiio I (1982) Methionine biosynthesis in Brevibacterium flavum: properties and essential role of O-acetylhomoserine sulfhydrylase. J Biochem 91:1163–1171
Parish T, Gordhan BG, McAdam RA, Duncan K, Mizrahi V, Stoker NG (1999) Production of mutants in amino acid biosynthesis genes of Mycobacterium tuberculosis by homologous recombination. Microbiology 145:3497–3503
Park S-D, Lee J-Y, Kim Y, Kim J-H, Lee H-S (1998) Isolation and analysis of metA, a methionine biosynthetic gene encoding homoserine acetyltransferase in Corynebacterium glutamicum. Mol Cells 8:286–294
Paszewski A (1993) Sulfur amino acid metabolism and its regulation in fungi: studies with Aspergillus nidulans. Acta Biochim Pol 40:445–449
Paszewski A, Prazmo W, Nadolska J, Regulski M (1984) Mutations affecting the sulphur assimilation pathway in Aspergillus nidulans: their effect on sulphur amino acid metabolism. J Gen Microbiol 130:1113–1121
Paszewski A, Brzywczy J, Natorff R (1994) Sulphur metabolism. Prog Ind Microbiol 29:299–319
Patte J-C (1996) Biosynthesis of threonine and lysine. In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger JE (eds) Escherichia coli and Salmonella: cellular and molecular biology, 2nd edn. ASM Press, Washington DC, pp. 528–541
Phillips SEV, Stockley PG (1996) Structure and function of Escherichia coli met repressor: similarities and contrasts with trp repressor. Philos Trans R Soc Lond B Biol Sci 351:527–35
Ravanel S, Droux M, Douce R (1995) Methionine biosynthesis in higher plants. I. Purification and characterization of cystathionine γ-synthase from spinach chloroplasts. Arch Biochem Biophys 316:572–584
Ron EZ (1975) Growth rate of Enterobacteriaceae at elevated temperatures: limitation by methionine. J Bacteriol 124:243-246
Ron EZ, Shani M (1971) Growth rate of Escherichia coli at elevated temperatures: reversible inhibition of homoserine trans-succinylase. J Bacteriol 107:397–400
Rossol I, Pühler A (1992) The Corynebacterium glutamicum aecD gene encodes a C-S lyase with α,β-elimination activity that degrades aminoethylcysteine. J Bacteriol 174:2968-2977
Rowbury, RJ (1983) Methionine biosynthesis and its regulation. In: Herman KM, Somervill RL (eds) Amino acids:biosynthesis and genetic regulation. Addison-Wesley, Reading, Massachusetts, pp. 191–211
Saint-Girons I, Parsot C, Zakin MM, Bârzu O, Cohen GN (1988) Methionine biosynthesis in Enterobacteriaceae: biochemical, regulatory, and evolutionary aspects. CRC Crit Rev Biochem 23(Suppl 1):S1—S42
Sahm H, Eggeling L, Eikmanns B, Krämer R (1995) Metabolic design in amino acid producing bacterium Corynebacterium glutamicum. FEMS Microbiol Rev 16:243–252
Sahm H, Eggeling L, de Graaf AA (2000) Pathway analysis and metabolic engineering in Corynebacterium glutamicum. Biol Chem 381:899-910
Schrumpf B, Schwarzer A, Kalinowski J, Pühler A, Eggeling L, Sahm H (1991) A functionally split pathway for lysine synthesis in Corynebacterium glutamicum. J Bacteriol 173:4510–4516
Sekowska A, Kung H-F, Danchin A (2000) Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J Mol Microbiol Biotechnol 2:145–177
Shiio I, Ozaki H (1981) Feedback inhibition by methionine and S-adenosylmethionine, and desensitization of homoserine O-acetyltransferase in Brevibacterium flavum. J Biochem 89:1493–1500
Shiio I, Ozaki H (1987) O-Acetylhomoserine sulfhydrylase from Brevibacterium flavum. Methods Enzymol 143:470–474
Shimizu H, Yamagata S, Masui R, Inoue Y, Shibata T, Yokoyama S, Kuramitsu S, Iwama T (2001) Cloning and overexpression of the oah1 gene encoding O-acetyl-l-homoserine sulfhydrylase of Thermus thermophilus HB8 and characterization of the gene product. Biochim Biophys Acta 1549:61–72
Simon M, Hong J-S (1983) Direct homocysteine biosynthesis from O-succinylhomoserine in Escherichia coli: an alternate pathway that bypasses cystathionine. J Bacteriol 153:558–561
Simonics T, Bánszky L, Maráz A (2002) Genetics of sulphate assimilation in Schizosaccharomyces pombe. Acta Microbiol Immunol Hung 49:279–283
Smith DA (1971) S-Amino acid metabolism and its regulation in Escherichia coli and Salmonella typhimurium. Adv Genet 16:141–165
Smith DA, Parish T, Stoker NG, Bancroft GJ (2001) Characterization of auxotrophic mutants of Mycobacterium tuberculosis and their potential as vaccine candidates. Infect Immun 69:1142–1150
Stipanuk MH (1986) Metabolism of sulfur-containing amino acids. Annu Rev Nutr 6:179–209
Taté R, Riccio A, Caputo E, Iaccarino M, Patriarca EJ (1999) The Rhizobium etli metZ gene is essential for methionine biosynthesis and nodulation of Phaseolus vulgaris. Mol Plant Microbe Interact 12:24–34
Thomas D, Surdin-Kerjan Y (1997) Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 61:503–532
Umbarger HE (1978) Amino acid biosynthesis and its regulation. Annu Rev Biochem 47:533–606
Urbanowski ML, Stauffer LT, Plamann LS, Stauffer GV (1987) A new methionine locus, metR, that encodes a trans-acting protein required for activation of metE and metH in Escherichia coli and Salmonella typhimurium. J Bacteriol 169:1391–1397
Vermeij P, Kertesz MA (1999) Pathways of assimilative sulfur metabolism in Pseudomonas putida. J Bacteriol 181:5833–5837
Wada M, Awano N, Haisa K, Takagi H, Nakamori S (2002) Purification, characterization and identification of cysteine desulfhydrase of Corynebacterium glutamicum, and its relationship to cysteine production. FEMS Microbiol Lett 217:103–107
Weissbach H, Brot N (1991) Regulation of methionine synthesis in Escherichia coli. Mol Microbiol 5:1593–1597
Wiebers JL, Garner HR (1967) Homocysteine and cysteine synthetases of Neurospora crassa. Purification, properties, and feedback control of activity. J Biol Chem 242:12–23
Wild CM, McNally T, Phillips SEV, Stockley PG (1996) Effects of systematic variation of the minimal Escherichia coli met consensus operator site: in vivo and in vitro met repressor binding. Mol Microbiol 21:1125–1135
Williams SJ, Senaratne RH, Mougous JD, Riley LW, Bertozzi CR (2002) 5′-Adenosinephosphosulfate lies at a metabolic branch point in mycobacteria. J Biol Chem 277:32606–32615
Wyman A, Paulus H (1975) Purification and properties of homoserine transacetylase from Bacillus polymyxa. J Biol Chem 250:3897–3903
Wyman A, Shelton E, Paulus H (1975) Regulation of homoserine transacetylase in whole cells of Bacillus polymyxa. J Biol Chem 250:3904–3908
Yamagata S (1989) Roles of O-acetyl-l-homoserine sulfhydrylases in microorganisms. Biochimie 71:1125–1143
Yamagata S, Ichioka K, Goto K, Mizuno Y, Iwama T (2001) Occurrence of transsulfuration in synthesis of l-homocysteine in an extremely thermophilic bacterium, Thermus thermophilus HB8. J Bacteriol 183:2086–2092
Acknowledgements
The researchers thank Dr. Younhee Kim and Mr. Matt Witherspoon for their careful reading and constructive comments on the manuscript. This work was supported by grants from BASF Korea (to H.-S. Lee) and the Ministry of Science and Technology (via 21C Microbial Genomics and Applications Center to H.-S. Lee).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, HS., Hwang, BJ. Methionine biosynthesis and its regulation in Corynebacterium glutamicum: parallel pathways of transsulfuration and direct sulfhydrylation. Appl Microbiol Biotechnol 62, 459–467 (2003). https://doi.org/10.1007/s00253-003-1306-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-003-1306-7