Abstract
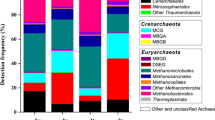
Mounting evidence suggests that Archaea are widespread and abundant in aquatic and terrestrial habitats and play fundamental roles in global biogeochemical cycles, yet the pattern and its ecological drivers of biogeographic distribution of archaeal community in estuarine ecosystem are still not well understood. Here, we investigated planktonic and benthic archaeal communities in the human-impacted Jiulong River estuary (JRE), southern China by using real-time PCR (RT-PCR) and Illumina 16S ribosomal RNA (rRNA) amplicon sequencing. RT-PCR analysis indicated that Archaea accounted for an average of 0.79 and 5.31 % of prokaryotic biomass in water and sediment samples of the JRE, respectively. The diversity of planktonic archaeal community decreased gradually from the river runoff to seawater, whereas that of benthic community did not show the similar pattern. The results of taxonomic assignments indicated that Thaumarchaeota (Nitrosopumilus and Cenarchaeum), Methanocorpusculum, and Methanospirillum were significantly more abundant in planktonic than benthic communities, whereas the relative abundances of Miscellaneous Crenarchaeotic Group, Marine Benthic Group-B/-D, anaerobic methane-oxidizing Archaea -1/-2D, and South Africa Gold Mine Euryarchaeotic Group 1 were higher in sediments than in surface waters. Moreover, planktonic archaeal community composition varied significantly at broad and finer-scale taxonomic levels along the salinity gradient. Multivariate statistical analyses revealed that salinity is the main factor structuring the JRE planktonic but not benthic archaeal community at both total community and population level. SourceTrakcer analysis indicated that river might be a major source of archaea in the freshwater zone of the JRE. Overall, this study advances our understanding of the biogeographic patterns and its ecological drivers of estuarine archaeal communities.







Similar content being viewed by others
References
Fuhrman JA (2009) Microbial community structure and its functional implications. Nature 459:193–199. doi:10.1038/nature08058
Dupont CL, Larsson J, Yooseph S, Ininbergs K, Goll J, Asplund-Samuelsson J, McCrow JP, Celepli N, Allen LZ, Ekman M, Lucas AJ, Hagstrom A, Thiagarajan M, Brindefalk B, Richter AR, Andersson AF, Tenney A, Lundin D, Tovchigrechko A, Nylander JAA, Brami D, Badger JH, Allen AE, Rusch DB, Hoffman J, Norrby E, Friedman R, Pinhassi J, Venter JC, Bergman B (2014) Functional tradeoffs underpin salinity-driven divergence in microbial community composition. PLoS One 9:e89549. doi:10.1371/journal.pone.0089549
Herlemann DPR, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ, Andersson AF (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579. doi:10.1038/ismej.2011.41
Fortunato CS, Eiler A, Herfort L, Needoba JA, Peterson TD, Crump BC (2013) Determining indicator taxa across spatial and seasonal gradients in the Columbia River coastal margin. ISME J 7:1899–1911. doi:10.1038/ismej.2013.79
Crump BC, Hopkinson CS, Sogin ML, Hobbie JE (2004) Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl Environ Microbiol 70:1494–1505. doi:10.1128/AEM. 70.3.1494-1505.2004
Stahl DA, de la Torre JR (2012) Physiology and diversity of ammonia-oxidizing archaea. Annu Rev Microbiol 66:83–101. doi:10.1146/annurev-micro-092611-150128
Offre P, Spang A, Schleper C (2013) Archaea in biogeochemical cycles. Annu Rev Microbiol 67:437–457. doi:10.1146/annurev-micro-092412-155614
Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P (2008) Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 6:245–252. doi:10.1038/nrmicro1852
Schleper C, Jurgens G, Jonuscheit M (2005) Genomic studies of uncultivated archaea. Nat Rev Microbiol 3:479–488. doi:10.1038/nrmicro1159
Karner MB, DeLong EF, Karl DM (2001) Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507–510. doi:10.1038/35054051
Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C (2003) Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol 5:787–797. doi:10.1046/j.1462-2920.2003.00476.x
Nicol GW, Schleper C (2006) Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol 14:207–212. doi:10.1016/j.tim.2006.03.004
Molari M, Giovannelli D, d’Errico G, Manini E (2012) Factors influencing prokaryotic community structure composition in sub-surface coastal sediments. Estuar Coast Shelf Sci 97:141–148. doi:10.1016/j.ecss.2011.11.036
Liu J, Yu S, Zhao M, He B, Zhang XH (2014) Shifts in archaeaplankton community structure along ecological gradients of Pearl Estuary. FEMS Microbiol Ecol. doi:10.1111/1574-6941.12404
Xie W, Zhang C, Zhou X, Wang P (2014) Salinity-dominated change in community structure and ecological function of Archaea from the lower Pearl River to coastal South China Sea. Appl Microbiol Biotechnol 98:7971–7982. doi:10.1007/s00253-014-5838-9
Crump BC, Baross JA (2000) Archaeaplankton in the Columbia River, its estuary and the adjacent coastal ocean, USA. FEMS Microbiol Ecol 31:231–239. doi:10.1111/j.1574-6941.2000.tb00688.x
Galand PE, Lovejoy C, Vincent WF (2006) Remarkably diverse and contrasting archaeal communities in a large arctic river and the coastal Arctic Ocean. Aquat Microb Ecol 44:115–126. doi:10.3354/ame044115
Galand PE, Lovejoy C, Pouliot J, Vincent WF (2008) Heterogeneous archaeal communities in the particle-rich environment of an arctic shelf ecosystem. J Mar Syst 74:774–782. doi:10.1016/j.jmarsys.2007.12.001
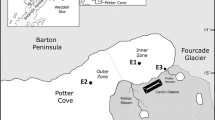
Li Q, Wang F, Chen Z, Yin X, Xiao X (2012) Stratified active archaeal communities in the sediments of Jiulong River estuary, China. Front Microbiol 3:311. doi:10.3389/fmicb.2012.00311
Yan XL, Zhai WD, Hong HS, Li Y, Guo WD, Huang X (2012) Distribution, fluxes and decadal changes of nutrients in the Jiulong River Estuary, Southwest Taiwan Strait. Chin Sci Bull 57:2307–2318. doi:10.1007/s11434-012-5084-4
Hu A, Yang X, Chen N, Hou L, Ma Y, Yu CP (2014) Response of bacterial communities to environmental changes in a mesoscale subtropical watershed, Southeast China. Sci Total Environ 472:746–756. doi:10.1016/j.scitotenv.2013.11.097
Guo W, Yang L, Hong H, Stedmon CA, Wang F, Xu J, Xie Y (2011) Assessing the dynamics of chromophoric dissolved organic matter in a subtropical estuary using parallel factor analysis. Mar Chem 124:125–133. doi:10.1016/j.marchem.2011.01.003
Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST (2011) Bayesian community-wide culture-independent microbial source tracking. Nat Meth 8:761–763. doi:10.1038/nmeth.1650
Cao WZ, Hong HS, Yue SP (2005) Modelling agricultural nitrogen contributions to the Jiulong River estuary and coastal water. Glob Planet Chang 47:111–121. doi:10.1016/j.gloplacha.2004.10.006
Yang YP, Hu MH (1997) The geochemistry of Jiulong River Estuary. In: Zhang J (ed) The biogeochemistry of major Chinese estuaries-transport of chemical materials and environment. Ocean Press, Beijing, pp 54–67
Zhai WD, Zhao HD, Zheng N, Xu Y (2012) Coastal acidification in summer bottom oxygen-depleted waters in northwestern-northern Bohai Sea from June to August in 2011. Chin Sci Bull 57:1062–1068. doi:10.1007/s11434-011-4949-2
Hu A, Jiao N, Zhang CL (2011) Community structure and function of planktonic Crenarchaeota: changes with depth in the South China Sea. Microb Ecol 62:549–563. doi:10.1007/s00248-011-9866-z
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Kemnitz D, Kolb S, Conrad R (2005) Phenotypic characterization of Rice Cluster III archaea without prior isolation by applying quantitative polymerase chain reaction to an enrichment culture. Environ Microbiol 7:553–565. doi:10.1111/j.1462-2920.2005.00723.x
Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C (2009) Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J 3:860–869. doi:10.1038/ismej.2009.23
Zhou HW, Li DF, Tam NFY, Jiang XT, Zhang H, Sheng HF, Qin J, Liu X, Zou F (2011) BIPES, a cost-effective high-throughput method for assessing microbial diversity. ISME J 5:741–749. doi:10.1038/ismej.2010.160
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi:10.1128/AEM. 01541-09
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Meth 7:335–336. doi:10.1038/nmeth.f.303
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi:10.1093/bioinformatics/btr381
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi:10.1093/bioinformatics/btq461
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Meth 10:996–998. doi:10.1038/nmeth.2604
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi:10.1186/gb-2011-12-6-r60
Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR (2011) Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS ONE 6:e16626. doi:10.1371/journal.pone.0016626
de la Torre JR, Walker CB, Ingalls AE, Konneke M, Stahl DA (2008) Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol 10:810–818. doi:10.1111/j.1462-2920.2007.01506.x
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M (2008) A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A 105:2134–2139. doi:10.1073/pnas.0708857105
Jung MY, Well R, Min D, Giesemann A, Park SJ, Kim JG, Kim SJ, Rhee SK (2014) Isotopic signatures of N2O produced by ammonia-oxidizing archaea from soils. ISME J 8:1115–1125. doi:10.1038/ismej.2013.205
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546. doi:10.1038/nature03911
Lebedeva EV, Hatzenpichler R, Pelletier E, Schuster N, Hauzmayer S, Bulaev A, Grigor’eva NV, Galushko A, Schmid M, Palatinszky M, Le Paslier D, Daims H, Wagner M (2013) Enrichment and genome sequence of the group I. 1a ammonia-oxidizing Archaeon “Ca. Nitrosotenuis uzonensis” representing a clade globally distributed in thermal habitats. PLoS One 8:e80835. doi:10.1371/journal.pone.0080835
Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW (2011) Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci U S A 108:15892–15897. doi:10.1073/pnas.1107196108
Mosier AC, Allen EE, Kim M, Ferriera S, Francis CA (2012) Genome sequence of “Candidatus Nitrosoarchaeum limnia” BG20, a low-salinity ammonia-oxidizing archaeon from the San Francisco Bay Estuary. J Bacteriol 194:2119–2120. doi:10.1128/JB.00007-12
Mosier AC, Allen EE, Kim M, Ferriera S, Francis CA (2012) Genome sequence of “Candidatus Nitrosopumilus salaria” BD31, an ammonia-oxidizing archaeon from the San Francisco Bay Estuary. J Bacteriol 194:2121–2122. doi:10.1128/JB.00013-12
Tourna M, Stieglmeier M, Spang A, Konneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108:8420–8425. doi:10.1073/pnas.1013488108
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N (2011) Examining the global distribution of dominant archaeal populations in soil. ISME J 5:908–917. doi:10.1038/ismej.2010.171
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi:10.1111/j.1442-9993.2001.01070.pp.x
Borcard D, Legendre P (2002) All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecol Model 153:51–68. doi:10.1016/S0304-3800(01)00501-4
Dray S, Legendre P, Peres-Neto PR (2006) Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecol Model 196:483–493. doi:10.1016/j.ecolmodel.2006.02.015
Blanchet FG, Legendre P, Borcard D (2008) Forward selection of explanatory variables. Ecology 89:2623–2632. doi:10.1890/07-0986.1
Oksanen J, Kindt R, Legendre P, O’Hara B, Simpson G, Solymos P, Stevens M, Wagner H (2009) Vegan: community ecology package. R package version 115-2
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi:10.1093/nar/gks1219
Wells LE, Cordray M, Bowerman S, Miller LA, Vincent WF, Deming JW (2006) Archaea in particle-rich waters of the Beaufort Shelf and Franklin Bay, Canadian Arctic: clues to an allochthonous origin? Limnol Oceanogr 51:47–59. doi:10.4319/lo.2006.51.1.0047
Hao DM, Tashiro T, Kato M, Sohrin R, Ishibashi T, Katsuyama C, Nagaosa K, Kimura H, Thanh TD, Kato K (2010) Population dynamics of Crenarchaeota and Euryarchaeota in the mixing front of river and marine waters. Microbes Environ 25:126–132
Auguet JC, Barberan A, Casamayor EO (2010) Global ecological patterns in uncultured Archaea. ISME J 4:182–190. doi:10.1038/ismej.2009.109
Campbell BJ, Kirchman DL (2013) Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME J 7:210–220. doi:10.1038/ismej.2012.93
Massana R, DeLong EF, Pedros-Alio C (2000) A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl Environ Microbiol 66:1777–1787. doi:10.1128/AEM. 66.5.1777-1787.2000
Zeng YH, Li HY, Jiao NZ (2007) Phylogenetic diversity of planktonic Archaea in the estuarine region of East China Sea. Microbiol Res 162:26–36. doi:10.1016/j.micres.2006.03.007
Kubo K, Lloyd KG, Biddle JF, Amann R, Teske A, Knittel K (2012) Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. ISME J 6:1949–1965. doi:10.1038/ismej.2012.37
Liu Y, Whitman WB (2008) Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann N Y Acad Sci 1125:171–189. doi:10.1196/annals.1419.019
Grossart HP, Frindte K, Dziallas C, Eckert W, Tang KW (2011) Microbial methane production in oxygenated water column of an oligotrophic lake. Proc Natl Acad Sci U S A 108:19657–19661. doi:10.1073/pnas.1110716108
Jurgens G, Glockner FO, Amann R, Saano A, Montonen L, Likolammi M, Munster U (2000) Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol Ecol 34:45–56. doi:10.1111/j.1574-6941.2000.tb00753.x
Auguet JC, Casamayor EO (2008) A hotspot for cold crenarchaeota in the neuston of high mountain lakes. Environ Microbiol 10:1080–1086. doi:10.1111/j.1462-2920.2007.01498.x
Herfort L, Kim JH, Coolen MJL, Abbas B, Schouten S, Herndl GJ, Damste JSS (2009) Diversity of Archaea and detection of crenarchaeotal amoA genes in the rivers Rhine and Têt. Aquat Microb Ecol 55:189–201. doi:10.3354/ame01294
Wang S, Dong RM, Dong CZ, Huang L, Jiang H, Wei Y, Feng L, Liu D, Yang G, Zhang C, Dong H (2012) Diversity of microbial plankton across the Three Gorges Dam of the Yangtze River, China. Geosci Front 3:335–349. doi:10.1016/j.gsf.2011.11.013
Vieira RP, Clementino MM, Cardoso AM, Oliveira DN, Albano RM, Gonzalez AM, Paranhos R, Martins OB (2007) Archaeal communities in a tropical estuarine ecosystem: Guanabara Bay, Brazil. Microb Ecol 54:460–468. doi:10.1007/s00248-007-9261-y
Pires ACC, Cleary DFR, Almeida A, Cunha A, Dealtry S, Mendonca-Hagler LCS, Smalla K, Gomes NCM (2012) Denaturing gradient Gel electrophoresis and barcoded pyrosequencing reveal unprecedented archaeal diversity in mangrove sediment and rhizosphere samples. Appl Environ Microbiol 78:5520–5528. doi:10.1128/AEM. 00386-12
Mosier AC, Francis CA (2008) Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ Microbiol 10:3002–3016. doi:10.1111/j.1462-2920.2008.01764.x
Beman JM, Francis CA (2006) Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl Environ Microbiol 72:7767–7777. doi:10.1128/AEM. 00946-06
Logares R, Brate J, Bertilsson S, Clasen JL, Shalchian-Tabrizi K, Rengefors K (2009) Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol 17:414–422. doi:10.1016/j.tim.2009.05.010
Acknowledgments
We thank Dr. Weidong Zhai and Ms. Xiuli Yan for providing physicochemical parameters of surface waters of the Jiulong River estuary and Mr. Jibing He and Mr. Jiajun Hong for assistance in collecting the samples. This work was supported by the National Natural Science Foundation of China (41106096), the Knowledge Innovation Program of the Chinese Academy of Sciences (IUEQN201307), Science and Technology Planning Project of Xiamen, China (3502Z20142021), and Natural Science Foundation of Ningbo, China (2013A610174, 2012C5011).
Conflict of Interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Anyi Hu got his Ph.D degree from Xiamen University.
Chang-Ping Yu got his Ph.D degree from National Taiwan University.
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Hu, A., Hou, L. & Yu, CP. Biogeography of Planktonic and Benthic Archaeal Communities in a Subtropical Eutrophic Estuary of China. Microb Ecol 70, 322–335 (2015). https://doi.org/10.1007/s00248-015-0597-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-015-0597-4