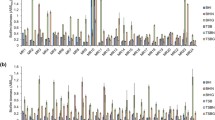
Abstract
Staphylococcus epidermidis has emerged as one of the major nosocomial pathogens associated with infections of implanted medical devices. The most important factor in the pathogenesis of these infections is the formation of bacterial biofilms. Bacteria grown in biofilms are more resistant to antibiotics and to the immune defence system than planktonic bacteria. In these infections, the antimicrobial therapy usually fails and the removal of the biofilm-coated implanted device is the only effective solution. In this study, three proteomic approaches were performed to investigate membrane proteins associated to biofilm formation: (i) sample fractionation by gel electrophoresis, followed by isotopic labelling and LC–MS/MS analysis, (ii) in-solution sample preparation, followed by isotopic labelling and LC–MS/MS analysis and (iii) in-solution sample preparation and label-free LC–MS/MS analysis. We found that the commensal strain S. epidermidis CECT 231 grown in biofilms expressed higher levels of five membrane and membrane-associated proteins involved in pathogenesis: accumulation-associated protein, staphylococcal secretory antigen, signal transduction protein TRAP, ribonuclease Y and phenol soluble modulin beta 1 when compared with bacteria grown under planktonic conditions. These results indicate that a commensal strain can acquire a pathogenic phenotype depending on the mode of growth.






Similar content being viewed by others
References
Anwar H, Costerton JW (1990) Enhanced activity of combination of tobramycin and piperacillin for eradication of sessile biofilm cells of Pseudomonas aeruginosa. Antimicrob Agents Chemother 34:1666–1671
Arber N, Pras E, Copperman Y, Schapiro JM, Meiner V, Lossos IS, Militianu A, Hassin D, Pras E, Shai A (1994) Pacemaker endocarditis. Report of 44 cases and review of the literature. Medicine (Baltimore) 73:299–305
Arciola CR, Campoccia D, Gamberini S, Donati ME, Pirini V, Visai L, Speziale P, Montanaro L (2005) Antibiotic resistance in exopolysaccharide-forming Staphylococcus epidermidis clinical isolates from orthopaedic implant infections. Biomaterials 26:6530–6535
Bjarnsholt T, Kirketerp-Moller K, Kristiansen S, Phipps R, Nielsen AK, Jensen PO, Hoiby N, Givskov M (2007) Silver against Pseudomonas aeruginosa biofilms. APMIS 115:921–928
Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc 4:484–494
Carroll J, Fearnley IM, Walker JE (2006) Definition of the mitochondrial proteome by measurement of molecular masses of membrane proteins. Proc Natl Acad Sci USA 103:16170–16175
Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322
Ehrlich GD, Hu FZ, Post JC (2004) Role for biofilms in infectious disease. In: Ghannoum M, O’Toole GA (eds) Microbial biofilms. ASM Press, Washington, DC, pp 332–358
Flemming HC, Wingender J (2010) The biofilm matrix. Nat Rev Microbiol 8:623–633
Hall-Stoodley L, Costerton JW, Stoodley P (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108
He X, Ahn J (2011) Differential gene expression in planktonic and biofilm cells of multiple antibiotic-resistant Salmonella typhimurium and Staphylococcus aureus. FEMS Microbiol Lett 325:180–188
Hopkins AL, Groom CR (2002) The druggable genome. Nat Rev Drug Discov 1:727–730
Hu J, Xu T, Zhu T et al (2011) Monoclonal antibodies against accumulation-associated protein affect EPS biosynthesis and enhance bacterial accumulation of Staphylococcus epidermidis. PLoS ONE 6:e20918
Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G (1997) A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun 65:519–524
Jones SM, Morgan M, Humphrey TJ, Lappin-Scott H (2001) Effect of vancomycin and rifampicin on meticillin-resistant Staphylococcus aureus biofilms. Lancet 357:40–41
Lang S, Livesley MA, Lambert PA, Littler WA, Elliott TS (2000) Identification of a novel antigen from Staphylococcus epidermidis. FEMS Immunol Med Microbiol 29:213–220
Li GZ, Vissers JP, Silva JC, Golick D, Gorenstein MV, Geromanos SJ (2009) Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9:1696–1719
Liduma I, Tracevska T, Bers U, Zilevica A (2012) Phenotypic and genetic analysis of biofilm formation by Staphylococcus epidermidis. Medicina (Kaunas) 48:305–309
Mack D, Becker P, Chatterjee I, Dobinsky S, Knobloch JK, Peters G, Rohde H, Herrmann M (2004) Mechanisms of biofilm formation in Staphylococcus epidermidis and Staphylococcus aureus: functional molecules, regulatory circuits, and adaptive responses. Int J Med Microbiol 294:203–212
Martin-Lopez JV, Diez-Gil O, Morales M, Batista N, Villar J, Claverie-Martin F, Mendez-Alvarez S (2004) Simultaneous PCR detection of ica cluster and methicillin and mupirocin resistance genes in catheter-isolated Staphylococcus. Int Microbiol 7:63–66
Molloy MP, Herbert BR, Slade MB, Rabilloud T, Nouwens AS, Williams KL, Gooley AA (2000) Proteomic analysis of the Escherichia coli outer membrane. Eur J Biochem 267:2871–2881
Oosthuizen MC, Steyn B, Theron J, Cosette P, Lindsay D, Von Holy A, Brozel VS (2002) Proteomic analysis reveals differential protein expression by Bacillus cereus during biofilm formation. Appl Environ Microbiol 68:2770–2780
Planchon S, Desvaux M, Chafsey I, Chambon C, Leroy S, Hebraud M, Talon R (2009) Comparative subproteome analyses of planktonic and sessile Staphylococcus xylosus C2a: new insight in cell physiology of a coagulase-negative Staphylococcus in biofilm. J Proteome Res 8:1797–1809
Resch A, Rosenstein R, Nerz C, Gotz F (2005) Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol 71:2663–2676
Resch A, Leicht S, Saric M, Pasztor L, Jakob A, Gotz F, Nordheim A (2006) Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 6:1867–1877
Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, Knobloch JK, Heilmann C, Herrmann M, Mack D (2005) Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol 55:1883–1895
Rupp ME, Ulphani JS, Fey PD, Mack D (1999a) Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect Immun 67:2656–2659
Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D (1999b) Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun 67:2627–2632
Sanchez CJ Jr, Mende K, Beckius ML, Akers KS, Romano DR, Wenke JC, Murray CK (2013) Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect Dis 13:47
Schumacher-Perdreau F, Heilmann C, Peters G, Gotz F, Pulverer G (1994) Comparative analysis of a biofilm-forming Staphylococcus epidermidis strain and its adhesion-positive, accumulation-negative mutant M7. FEMS Microbiol Lett 117:71–78
Schwank S, Rajacic Z, Zimmerli W, Blaser J (1998) Impact of bacterial biofilm formation on in vitro and in vivo activities of antibiotics. Antimicrob Agents Chemother 42:895–898
Seyer D, Cosette P, Siroy A, Dé E, Lenz C, Vaudry H, Coquet L, Jouenne T (2005) Proteomic comparison of outer membrane protein patterns of sessile and planktonic Pseudomonas aeruginosa cells. Biofilms 2:27–36
Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68:850–858
Shin JH, Lee HW, Kim SM, Kim J (2009) Proteomic analysis of Acinetobacter baumannii in biofilm and planktonic growth mode. J Microbiol 47:728–735
Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S (2005) Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem 77:2187–2200
Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics 5:144–156
Siroy A, Cosette P, Seyer D, Lemaître-Guillier C, Vallenet D, Van Dorsselaer A, Boyer-Mariotte S, Jouenne T, Dé E (2006) Global comparison of the membrane subproteomes between a multidrug-resistant Acinetobacter baumannii strain and a reference strain. J Proteome Res 5:3385–3398
Soares NC, Cabral MP, Parreira JR, Gayoso C, Barba MJ, Bou G (2009) 2-DE analysis indicates that Acinetobacter baumannii displays a robust and versatile metabolism. Proteome Sci 7:37
Stevens NT, Greene CM, O’Gara JP, Humphreys H (2009) Biofilm characteristics of Staphylococcus epidermidis isolates associated with device-related meningitis. J Med Microbiol 58:855–862
Steyn B, Oosthuizen MC, MacDonald R, Theron J, Brozel VS (2001) The use of glass wool as an attachment surface for studying phenotypic changes in Pseudomonas aeruginosa biofilms by two-dimensional gel electrophoresis. Proteomics 1:871–879
Sun D, Accavitti MA, Bryers JD (2005) Inhibition of biofilm formation by monoclonal antibodies against Staphylococcus epidermidis RP62A accumulation-associated protein. Clin Diagn Lab Immunol 12:93–100
Szczuka E, Kaznowski A (2014) Antimicrobial activity of tigecycline alone or in combination with rifampin against Staphylococcus epidermidis in biofilm. Folia Microbiol 59:283–288
Szczuka E, Urbanska K, Pietryka M, Kaznowski A (2013) Biofilm density and detection of biofilm-producing genes in methicillin-resistant Staphylococcus aureus strains. Folia Microbiol (Praha) 58:47–52
Tan S, Tan HT, Chung MC (2008) Membrane proteins and membrane proteomics. Proteomics 8:3924–3932
van Alen T, Claus H, Zahedi RP, Groh J, Blazyca H, Lappann M, Sickmann A, Vogel U (2010) Comparative proteomic analysis of biofilm and planktonic cells of Neisseria meningitidis. Proteomics 10:4512–4521
Visutthi M, Srimanote P, Voravuthikunchai SP (2011) Responses in the expression of extracellular proteins in methicillin-resistant Staphylococcus aureus treated with rhodomyrtone. J Microbiol 49:956–964
Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362
World Health Organization (2004) Laboratory biosafety manual, 3rd edn. WHO, Geneva
Yan L, Zhang L, Ma H, Chiu D, Bryers JD (2014) A single B-repeat of Staphylococcus epidermidis accumulation-associated protein induces protective immune responses in an experimental biomaterial-associated infection mouse model. Clin Vaccine Immunol 21:1206–1214
Yang XM, Li N, Chen JM, Ou YZ, Jin H, Lu HJ, Zhu YL, Qin ZQ, Qu D, Yang PY (2006) Comparative proteomic analysis between the invasive and commensal strains of Staphylococcus epidermidis. FEMS Microbiol Lett 261:32–40
Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM (2003) Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol Microbiol 49:1577–1593
Acknowledgments
S.A-A thanks the Basque Government and the Medical Research Council for supporting her stage in Cambridge. We also thank Eneritz Bilbao and Joe Carroll for their excellent technical assistance. The authors acknowledge the Analytical and High-Resolution Microscopy in Biomedicine Service at the University of Basque Country for assistance with Scanning Electron Microscopy. The Proteomics Core Facility-SGIKER (member of ProteoRed-ISCIII) is supported by Universidad del País Vasco/Euskal Herriko Unibertsitatea, Ministerio de Ciencia e Innovación, Gobierno Vasco/Eusko Jaurlaritza, and European Science Foundation. This work was supported by funds from the Spanish Ministry of Economy (Grant No. BFU2012-36241) and from MICINN (Grant No. BFU2010-22103). S.A-A was a pre-doctoral student supported by the Basque Government and by the Fundación Biofísica Bizkaia. We thank Prof. Holger Rohde from the Institut für Medizinische Mikrobiologie, Virologie und Hygiene (Hamburg, Germany) for supplying the anti-Aap antiserum.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Águila-Arcos, S., Ding, S., Aloria, K. et al. A Commensal Strain of Staphylococcus epidermidis Overexpresses Membrane Proteins Associated with Pathogenesis When Grown in Biofilms. J Membrane Biol 248, 431–442 (2015). https://doi.org/10.1007/s00232-015-9801-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00232-015-9801-1