Abstract
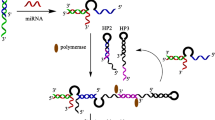
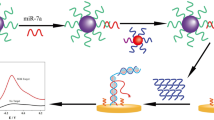
Infectious diseases are a long-standing and severe global public health problem. The rapid diagnosis of infectious diseases is an urgent need to solve this problem. MicroRNA (miRNA) plays an important role in the intervention of some infectious diseases and is expected to become a potential biomarker for the diagnosis and prognosis of infectious diseases. It is of great significance to develop rapid and sensitive methods for detecting miRNA for effective control of infectious diseases. In this study, a simple and highly sensitive homogeneous electrochemical method for microRNAs using enzyme-driven cascaded signal amplification has been developed. In the presence of target miRNA, the reaction system produced plenty of MB-labeled single-nucleotide fragments (MB-MF) containing a few negative charges, which can diffuse to the negative surface of the ITO electrode easily, so an obvious electrochemical signal enhancement was obtained. Without the target, MB-HP contains a relatively large amount of negative charges due to the phosphates on the DNA chain, which cannot be digested by the enzyme and cannot diffuse freely to the negatively charged ITO electrode, so only a small signal was detected. The enhanced electrochemical response has a linear relationship with the logarithm of miRNA concentration in the range of 10 fM to 10 nM and the limit of detection as low as 3.0 fM. Furthermore, the proposed strategy showed the capability of discriminating single-base mismatch and performed eligibly in the analysis of miRNA in cell lysates, exhibiting great potential for disease diagnosis and biomedical research.

Graphical abstract





Similar content being viewed by others
References
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8.
Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285(2):116–26.
Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13(5):358–69.
Tricoli JV, Jacobson JW. MicroRNA: potential for cancer detection, diagnosis, and prognosis. Cancer Res. 2007;67(10):4553.
Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10(6):389–402.
Saito M, Schetter AJ, Mollerup S, Kohno T, Skaug V, Bowman ED, et al. The association of microRNA expression with prognosis and progression in early-stage, non–small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17(7):1875.
Swaminathan G, Rossi F, Sierra L-J, Gupta A, Navas-Martin S, Martin-Garcia J. A role for microRNA-155 modulation in the anti-HIV-1 effects of toll-like receptor 3 stimulation in macrophages. PLoS Pathog. 2012;8(9):e1002937.
Natekar JP, Rothan HA, Arora K, Strate PG, Kumar M. Cellular microRNA-155 regulates virus-induced inflammatory response and protects against lethal West Nile virus infection. Viruses. 2020;12(1).
Liu S, Lin Y, Wang L, Liu T, Cheng C, Wei W, et al. Exonuclease III-aided autocatalytic DNA biosensing platform for immobilization-free and ultrasensitive electrochemical detection of nucleic acid and protein. Anal Chem. 2014;86(8):4008–15.
Hu T, Zhang L, Wen W, Zhang X, Wang S. Enzyme catalytic amplification of miRNA-155 detection with graphene quantum dot-based electrochemical biosensor. Biosens Bioelectron. 2016;77:451–6.
Zeng D, Wang Z, Meng Z, Wang P, San L, Wang W, et al. DNA tetrahedral nanostructure-based electrochemical miRNA biosensor for simultaneous detection of multiple miRNAs in pancreatic carcinoma. ACS Appl Mater Interfaces. 2017;9(28):24118–25.
Tran HV, Piro B, Reisberg S, Huy Nguyen L, Dung Nguyen T, Duc HT, et al. An electrochemical ELISA-like immunosensor for miRNAs detection based on screen-printed gold electrodes modified with reduced graphene oxide and carbon nanotubes. Biosens Bioelectron. 2014;62:25–30.
Zhang J, Hun X. Electrochemical determination of miRNA-155 using molybdenum carbide nanosheets and colloidal gold modified electrode coupled with mismatched catalytic hairpin assembly strategy. Microchem J. 2019;150:104095.
Duan F, Guo C, Hu M, Song Y, Wang M, He L, et al. Construction of the 0D/2D heterojunction of Ti3C2Tx MXene nanosheets and iron phthalocyanine quantum dots for the impedimetric aptasensing of microRNA-155. Sens. Actuators, B. 2020;310:127844.
Xu S, Chang Y, Wu Z, Li Y, Yuan R, Chai Y. One DNA circle capture probe with multiple target recognition domains for simultaneous electrochemical detection of miRNA-21 and miRNA-155. Biosens Bioelectron. 2020;149:111848.
Liu L, Zhu S, Wei Y, Liu X, Jiao S, Yang J. Ultrasensitive detection of miRNA-155 based on controlled fabrication of AuNPs@MoS2 nanostructures by atomic layer deposition. Biosens Bioelectron. 2019;144:111660.
Wang X, Jiang A, Hou T, Li F. A sensitive and versatile “signal-on” electrochemical aptasensor based on a triple-helix molecular switch. Analyst. 2014;139(23):6272–8.
Zhou L, Ou L-J, Chu X, Shen G-L, Yu R-Q. Aptamer-based rolling circle amplification: a platform for electrochemical detection of protein. Anal Chem. 2007;79(19):7492–500.
Liu A, Wang K, Weng S, Lei Y, Lin L, Chen W, et al. Development of electrochemical DNA biosensors. Trac-trend Anal Chem. 2012;37:101–11.
Xuan F, Luo X, Hsing IM. Ultrasensitive solution-phase electrochemical molecular beacon-based DNA detection with signal amplification by exonuclease III-assisted target recycling. Anal Chem. 2012;84(12):5216–20.
Luo X, Lee TM-H, Hsing IM. Immobilization-free sequence-specific electrochemical detection of DNA using ferrocene-labeled peptide nucleic acid. Anal Chem. 2008;80(19):7341–6.
Zhang Y, Xia J, Zhang F, Wang Z, Liu Q. A dual-channel homogeneous aptasensor combining colorimetric with electrochemical strategy for thrombin. Biosens Bioelectron. 2018;120:15–21.
Liu F, Yang L, Yin X, Liu X, Ge L, Li F. A facile homogeneous electrochemical biosensing strategy based on displacement reaction for intracellular and extracellular hydrogen peroxide detection. Biosens Bioelectron. 2019;141:111446.
Fu C, Liu C, Li Y, Guo Y, Luo F, Wang P, et al. Homogeneous electrochemical biosensor for melamine based on DNA triplex structure and exonuclease III-assisted recycling amplification. Anal Chem. 2016;88:10176–82.
Hou T, Li W, Liu X, Li F. Label-free and enzyme-free homogeneous electrochemical biosensing strategy based on hybridization chain reaction: a facile, sensitive, and highly specific MicroRNA assay. Anal Chem. 2015;87(22):11368–74.
Fu C, Liu C, Wang S, Luo F, Lin Z, Chen G. A signal-on homogeneous electrochemical biosensor for sequence-specific microRNA based on duplex-specific nuclease-assisted target recycling amplification. Anal Methods. 2016;8(39):7034–9.
Zhou W, Gong X, Xiang Y, Yuan R, Chai Y. Quadratic recycling amplification for label-free and sensitive visual detection of HIV DNA. Biosens Bioelectron. 2014;55:220–4.
Liu SF, Wang CF, Zhang CX, Wang Y, Tang B. Label-free and ultrasensitive electrochemical detection of nucleic acids based on autocatalytic and exonuclease III-assisted target recycling strategy. Anal Chem. 2013;85(4):2282–8.
Wang L-j, Ren M, Zhang Q, Tang B, C-y Z. Excision repair-initiated enzyme-assisted bicyclic cascade signal amplification for ultrasensitive detection of uracil-DNA glycosylase. Anal Chem. 2017;89(8):4488–94.
Xiong E, Yan X, Zhang X, Liu Y, Zhou J, Chen J. Exonuclease III-assisted cascade signal amplification strategy for label-free and ultrasensitive electrochemical detection of nucleic acids. Biosens Bioelectron. 2017;87:732–630.
Song X, Hou T, Lu F, Wang Y, Liu J, Li F. Homogeneous photoelectrochemical biosensing via synergy of G-quadruplex/hemin catalysed reactions and the inner filter effect. Chem Commun. 2020;56(12):1811–4.
Miao X, Cheng Z, Ma H, Li Z, Xue N, Wang P. Label-free platform for microRNA detection based on the fluorescence quenching of positively charged gold nanoparticles to silver nanoclusters. Anal Chem. 2018;90(2):1098–103.
Borghei YS, Hosseini M, Ganjali MR. Oxidase-like catalytic activity of Cys-AuNCs upon visible light irradiation and its application for visual miRNA detection. Sens. Actuators, B 2018;273:1618–1626.
Funding
This project was financially supported by the National Sciences Foundation of China (21775026, 21974020); the cooperative project of production and study in the University of Fujian Province (2018Y4007); and the Sciences Foundation of Fujian Province (2018J01685, 2018J01682).
Author information
Authors and Affiliations
Contributions
The manuscript was written through contributions of all authors. C.Lin and Z.Lin conceived the projects, and Y. Huang, X. Huang, and H.Z. designed and performed the experiments and collected the data. Y. Huang, X. Huang, H. Zheng, and Z. Lin analyzed and discussed the data. All authors discussed the results and contributed to the writing of this manuscript. All authors have given approval to the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Published in the topical collection Analytical Chemistry for Infectious Disease Detection and Prevention with guest editors Chaoyong Yang and XiuJun (James) Li.
Rights and permissions
About this article
Cite this article
Huang, Y., Huang, X., Zheng, H. et al. Homogeneous electrochemical biosensor for microRNA based on enzyme-driven cascaded signal amplification strategy. Anal Bioanal Chem 413, 4681–4688 (2021). https://doi.org/10.1007/s00216-020-03027-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-03027-3