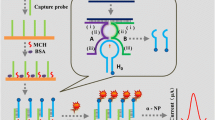
Abstract
The utility of DNA microarrays is severely limited by their restricted sensitivity. Tyramine signal amplification (TSA) coupled with gold label silver stain (GLSS) was introduced in DNA microarrays for visual detection of human pathogenic microorganisms. First, a TSA system was introduced to the microarrays after the microarrays were prepared and hybridized with biotinylated targets. This procedure leads to large amounts of biotin-conjugated tyramine depositing at the site of enzyme reaction under HRP catalysis. Second, streptavidin–nanogold was introduced and accumulated by specific binding of biotin and streptavidin. Finally, silver staining was performed. The images of the spots were scanned with a visible light scanner and quantified with ArrayVision 7.0 software. Detection conditions were systematically optimized. Then the sensitivity among TSA coupled with GLSS, GLSS, and TSA coupled with Cy3 was compared. The optimized conditions were: streptavidin–HRP (1 mg mL−1) dilution 1:1500, biotin–tyramine dilution 1:200 (+0.5% H2O2), streptavidin–nanogold dilution 1:100 (all diluted in 1 × PBS + 1% BSA) and silver stain time of 10 min. The sensitivity of TSA coupled with GLSS was 100-fold higher than that of GLSS, and was identical with that of TSA coupled with Cy3. Meanwhile, the specificity of the microarrays were not affected. This implied that TSA coupled with GLSS was a sensitive visual detection method and would be an ideal alternative to fluorescence-based detection for DNA microarrays.

Scanned images and quantification of the microarrays in comparison of the sensitivity between TSA–GLSS and GLSS. The result showed that the sensitivity of this method was 100-fold higher than that of GLSS




Similar content being viewed by others
References
Schena M (1996) Genome analysis with gene expression microarrays. BioEssays 18:427–431
Brown PO, Botsein D (1999) Exploring the world of the genome with DNA microarrays. Nat Genet 21:33–37
Heller Renu A, Schena M, Chai A, Shalone D, Bedilion T, Gilmore J, Woolley D, Davis RW (1997) Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc Natl Acad Sci USA 94:2150–2155
Da-Zhi J, Si-Yuan W, Su-Hong C, Feng L, Sheng-Qi W (2006) Detection and identification of intestinal pathogens in clinical specimens using DNA microarrays. Mol Cell Probes 20:337–347
Schena M, Renu A (1998) Microarrays: biotechnology’s discovery platform for functional genomics. Tib Tech 16:301–306
Bowtelld DL (1999) Options available from start to finish-for obtaining expression data by microarray. Nat Genet 21:25–32
Rajeevan Mangalathu S, Dimulescu IM, Unger ER, Vernon SD (1999) Chemiluminescent analysis of gene expression on high-density filter arrays. J Histochem Cytochem 47:337–342
Stratis-Cullum DN, Griffin GD, Mobley J, Vo-Dinh T (2008) Intensified biochip system using chemiluminescence for the detection of Bacillus globigii spores. Anal Bioanal Chem 391:1655–1660
Alexandre I, Hamels S et al (2001) Visual silver detection of DNA microarrays. Anal Biochem 295:1–8
Ji M, Hou P, Li S, He N, Zuhong Lu (2004) Visual silver detection of methylation using DNA microarray coupled with linker-PCR. Clinica Chimica Acta 342:145–153
Cao X, Wang Y-F, Zhang C-F, Gao W-J (2006) Visual DNA microarrays for simultaneous detection of Ureaplasma urealyticum and Chlamydia trachomatis coupled with multiplex asymmetrical PCR. Biosens Bioelectron 22(3):393–398
Call DR, Borucki MK, Loge FJ (2003) Detection of bacterial pathogens in environmental samples using DNA microarrays. J Microbiol Methods 53:235–243
Bobrow MN, Harris TD, Shaughnessy KJ, Litt GJ (1989) Catalyzed reporter deposition, a novel method of signal amplification. Application to immunoassays. J Immunol Methods 125:279–285
Kerstens HMJ, Poddighe PJ, Hanselaar AGJM (1995) A novel in situ hybridization signal amplification method based on the deposition of biotinylated tyramine. J Histochem Cytochem 43:347–352
Mayer G, Bendayan M (1997) Biotinyl–tyramide: a novel approach for electron microscopic immunocytochemistry. J Histochem Cytochem 45:1449–1454
Seung-won L, Eun LS, Hyuk Ko Seong (2005) Introduction of Tyramide Signal Amplification (TSA) to pre-embedding nanogold–silver staining at the electron microscopic level. J Histochem Cytochem 53(2):249–252
Jin D, Qi H, Chen S, Zeng T, Liu Q, Wang S (2008) Simultaneous detection of six human diarrheal pathogens by using DNA microarray combined with tyramide signal amplification. J Microbiol Methods 75:365–368
Hopman AHN, Ramaekers FCS, Speel EJM (1998) Rapid synthesis of biotin-, digoxigenin-, trinitrophenyl-, and fluorochrome-labeled tyramides and their application for in situ hybridization using CARD amplification. J Histochem Cytochem 46(6):771–777
Zaitsu K, Ohkura Y (1980) New fluorogenic substrates for horseradish peroxidase: rapid and sensitive assays for hydrogen peroxide and the peroxidase. Anal Biochem 109:109–113
Speel EJM, Anton HN (1999) Amplification methods to increase the sensitivity of in situ hybridization: play CARD(S). J Histochem Cytochem 47(3):281–288
Acknowledgements
This work was supported by the National High-Tech R&D Program (863 Program) (2007AA02Z400) and the National science and technology Major project (2009ZX10004–310).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Qi, HJ., Chen, SH., Zhang, ML. et al. DNA microarrays for visual detection of human pathogenic microorganisms based on tyramine signal amplification coupled with gold label silver stain. Anal Bioanal Chem 398, 2745–2750 (2010). https://doi.org/10.1007/s00216-010-4189-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-010-4189-3