Abstract
Acrylates are used in cosmetics, orthopedics, paints, coatings, adhesives, textiles, and biomedical applications such as contact lenses and bone cements. However, some acrylates are mutagenic and the aim of this article is to explain the mutagenicity in terms of the atomic population redistribution in the molecule using two new descriptors which are based on atomic populations framed in the quantum theory of atoms in molecules. They describe the electron-withdrawing effect of a group of atoms in a molecule. The descriptors consider substituents of prop-2-enoates, the number of the acrolein units and the electrophilicity. The cluster analysis using these descriptors allows to classify acrylates in terms of the number of acrolein backbones and the type of the substituent group. Five main groups can be distinguished: monoacrylates with monomethacrylates, diacrylates with dimethacrylates, triacrylates, trimethacrylate and monoacrylates with electron-rich substituents. The substituents of mutagenic acrylates are electron withdrawing. This makes the acrolein backbone β-carbon more electrophilic and the molecule more reactive.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Acrylates are the esters of acrylic acid. Their IUPAC name is prop-2-enoates and they are extensively used in the industry [1]. Despite their extensive use, there are few reported studies related to a prediction of their properties. Yu et al. proposed an artificial neural network (ANN) model to predict the reactivity of 34 acrylate monomers in radical copolymerization. This model was build on quantum–mechanical descriptors such as Mulliken charges and highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) energies [2]. In a study conducted by Perez-Garrido et al, a quantitative structure–activity relationship (QSAR) model was proposed to predict the mutagenicity of a set of more than 100 acrylates [3]. This model was build on the basis of more than 1000 descriptors encoded in DRAGON software [4]. A study of Tanis et al., focused on the chemical reactivity of the N-(4-nitrophenyl)acrylamide (N4PA) with nucleic acids. This reactivity was studied with the quantum mechanical descriptors such as the electronic energy, the electronegativity, the chemical hardness, the chemical potential and HOMO–LUMO energies [5]. Furuhama et al. studied acrylates in order to predict their ecotoxicity employing a QSAR model and using Gasteiger’s partial equalization of orbital electronegativity (PEOE) as descriptors [6]. To determine the toxicity of methacrylates Ishihara et al. used quantum chemical descriptors such as HOMO, LUMO, electronegativity and partition coefficient (logP) in a QSAR model. They found that their model may help them to reveal the toxic potentials of new medical and dental materials, thereby facilitating the synthesis of less toxic acrylates [7]. These authors also estimated the hemolytic activity of aliphatic and aromatic methacrylates using the following quantum chemical descriptors such as electron affinity, ionization potential, HOMO and LUMO energies, and the partition coefficient (logP) [8]. Liu et al studied a set of polymethacrylates to reproduce physical properties such as molar volume at room temperature, refractive index and glass transition temperature using descriptors such as the side chain length, polarizability and HOMO energy. Their model predicted reasonably well the properties of acrylate polymers [9]. Acrylates have also been classified using descriptors such as the number of acrylate groups, the calculated value of LogP, and the number of molecular paths [10]. In this study, 143 acrylates were clustered in five groups, differing in their LogP values, their halogenation and their degree of branching.
No descriptor has been proposed so far to understand the atomic population redistribution of acrylates upon the interchange of ester substituents and the relation with their mutagenicity. Therefore, we propose two descriptors based on the framework of the quantum theory of atoms in molecules (QTAIM)[11,12,13,14]. The motivation for the development of these descriptors was to find out how substituent groups (ester group and substituent in α position) affect the atomic population of the acrolein backbone, and whether this allows to distinguish between different types of acrylates. The proposed descriptors are based on (i) the proportion between the QTAIM atomic population of the fragments and the acrolein backbone and (ii) the concept of electronegativity and electron affinity. QTAIM based descriptors have been proposed previously, however, the focus in that study was on electron delocalization [15]. The present work is focused on the atomic population redistribution upon substituent interchange on acrylates and its relation with the mutagenicity as the capacity to induce mutations and cancer [16]. Mutagenicity can be related to the Michael reactivity [17].
This paper contains the following sections: (i.) Atomic populations and descriptors, the relation between the electron density redistribution upon the addition of one electron and the values of both descriptors are presented. (ii.) Effects of the substituent in X position. (iii.) Effects of the substituent in Y position and (iv.) Hierarchical clustering based on the proposed descriptors.
2 Methodology
2.1 The selection of the molecules
The U.S. Environmental Protection Agency and the International Agency of Research on Cancer has classified some acrylates as a possible human carcinogens. For example, the exposure of some acrylates to human body tissues such as skin, throat or eye have been reported to induce serious health consequences such as reproductive toxicity, cancer, neurological damage, organ system toxicity and cellular damage [1]. The main interest in this study is to propose two descriptors describing the electron distribution in acrylates and its relation with mutagenicity. Therefore, we selected 65 acrylates from various databases [18,19,20] and references [21,22,23,24,25,26,27,28] of which 8 are mutagenic [18, 21, 22]. Among the 65 acrylates there are 25 acrylates, 19 methacrylates, 8 diacrylates, 10 dimethacrylates, 2 triacrylates and 1 trimethacrylate. The reported mutagenicity (Table 1) was evaluated with the Ames Test using the Salmonella typhimurium TA100 strain assays.
2.2 Structure of acrylates
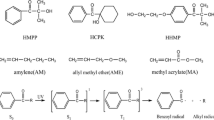
Acrylates are α,β-unsaturated carbonyl compounds, and may have different substituent groups in positions X and Y as shown in Scheme 1.
The mesomeric effect described in terms of electron withdrawal depends on the strength of an atom in a molecule to attract electrons, called electronegativity by Linus Pauling [32]. This concept can be extended to groups of atoms, functional groups, or substituents to attract electrons from their molecular counterpart. Based on this concept, we propose the two QTAIM descriptors in this article.
2.3 Computational methods
A conformational study was performed for each acrylate to select the most stable conformer. This study was done with the Conformer–Rotamer Ensemble Sampling Tool (CREST) [33]. The conformational study was performed with the GFN2-xTB [34] function with an energy threshold of 10 kcal/mol.
The most stable conformer was selected and reoptimized with the M06-2X [35] density functional and the 6-311G(d,p) valence double zeta basis set [36]. Atom pairwise dispersion corrections with zero damping were taken into account [37]. All calculations were performed with the ORCA quantum chemistry program package version 5.0 [38, 39]. The geometries of the most stable conformer for each acrylate have in common that the orientation of the double bond with the carbonyl bond is synperiplanar [40]. For each minimized geometry the wave function was obtained and analyzed using QTAIM [11, 12] with the AIMALL software [41].
This analysis allows to obtain the atomic basins and their corresponding properties, such as the atomic population, N(Ω). Integration errors were estimated through differences between molecular properties and those obtained by the summation of N(Ω). Their absolute values were always smaller than 2.00 × 10–3 a.u.
2.4 QTAIM based descriptors
We propose an arithmetic proportion that relates the electron-withdrawing effect of a group of atoms in a molecule. QTAIM allows partitioning (Eq. 1) of any molecular property P of a specific molecular system S into their atomic contributions P(Ω) where Ω denotes an atom in the system [11,12,13,14].
An application of this partitioning is the molecular population N which can be split into atomic contributions N(Ω). The electronegativity (EN) of substituent groups in an acrylate (Eq. 2) can be related to the quotient of atom populations N(Ω) between the atoms of substituent groups and those of the acrolein backbone A.
The electron affinity (EA) is the variation of the energy upon the addition of an electron to a neutral atom or molecule in the gas phase [42].
Within the QTAIM framework, the electron affinity can be decomposed into atomic contributions, denoted by Ω:
The variation of atomic population N(Ω) upon addition of an electron can be written in terms of the EA:
EN(Ω) and EA(Ω) can be merged into one equation, leading to the definition of the molecular descriptor D:
The external sum in Eq. 6 runs over all acrolein groups (R is the total number of acrolein substituents). In the case of i-th acrolein backbone, the numerator indicates the sum of atomic population differences of those atoms that do not belong to this structure. Among those atoms, it includes the remaining atoms of all other acrolein backbones and substituent fragments (Fig. 1). The denominator is the sum over the atomic population differences of all atoms that belong to i-th acrolein backbone.
In Eq. 7, we define the descriptor D∩. The numerator of this descriptor is the sum of the atomic population difference between the acrylate anion and the neutral acrylate of the atoms that do not belong to the acrolein backbone and the denominator is the sum of the same difference, but for the acrolein backbone atoms (Fig. 2). In the case of a molecule with two acrolein backbones and from the definition of D, this descriptor can be understood as the intersection of the first fragment with the second fragment (Fig. 1). All fragments that neither belong to the first fragment nor the second fragment are the substituent groups in positions X and Y in each acrolein backbone. Therefore, the intersected fragments are the acrolein backbones.
In the following, we will discuss the differences between the two descriptors in more detail. The aim of defining two descriptors is to understand how branched an acrylate is and how the atomic population changes depending on the substituents in X and Y positions (Scheme 1). The descriptor D indicates how branched the molecule is. Larger values of this descriptor correspond to a branched molecule with several acrolein backbones, while smaller values indicate that the molecule is not branched and has only one acrolein backbone. Larger values of the D∩ descriptor correspond to larger atomic population on the substituents.
2.5 Clustering analysis
A correlation analysis revealed that D and D∩ are not correlated (correlation coefficient of -0.038). In order to test the performance of the proposed descriptors, we applied them to a series of α-substituted methyl acrylates with the following substituent groups: –F, –Cl, –H, –CN, –CH3, –NO2. These acrylates were previously studied by Giraldo et al [43]. To this end, the obtained values of descriptors D and D∩ for the 65 acrylates were normalized using Eq. 8:
where X is the referenced value, Min and Max are the minimum and maximum values and X′ is the normalized value between [0, 1]. Using this normalized data, we carried out a cluster analysis using five different distance definitions: 1. Euclidean, 2. Manhattan, 3. Minkowski, 4. Maximum, and 5. Canberra and eight grouping methodologies: i. Single, ii. Complete linkage, iii. Average, iv. McQuitty, v. Centroid, vi. Ward.D, vii. Ward.D2 and viii. Median [44,45,46]. We obtained 40 dendrograms, eight for each similarity function, using the software R [47] and the dendextend package [48]. The best dendrogram was selected based on the cophenetic correlation [49], which was obtained with the Minkowski distance definition and the average grouping methodology.
3 Results and discussion
3.1 Atomic populations and descriptors
D and D∩ are introduced to understand the variation of the atomic population N(Ω) upon addition of one electron, and how it is affected by the substituents in X and Y positions (Scheme 1). The structures of all molecules studied herein and their name, numeration and mutagenicity are shown in Table 1.
Methyl acrylate (compound 1 in Table 1 and in Table S1) and ethyl acrylate (compound 47 in Table 1 and Table S1), both with one acrolein backbone, are taken as an example. As can be seen in Table 1, 75.9% of the electron are in the acrolein backbone for compound 1 and 75.2% for compound 47, while the rest is distributed on the substituents in X and Y positions (Scheme 1). Therefore, larger values of the D and D∩ descriptors (Table 2) indicate a less localized atomic population at the acrolein backbone.
Both descriptor values, D and D∩, depend on substituents in X and Y position, and on the number of acrolein backbones in the molecule. However, the value of D is much more strongly affected by the number of acrolein backbones. For example, the value of D in compound 6 (which is a diacrylate) is larger than in compound 3 (a monoacrylate) (Table 2). On the other hand, D∩ for compound 6 indicates that the added electron is more localized at the substituents in X and Y than for compound 3 (Table 2). The magnitude of these values reflects that the added electron in compound 3 is mainly located at the acrolein backbones rather than on the substituents (Eqs. 6 and 7).
3.2 Effects of the substituent in X position
We now analyze in more detail the effect of the substituent in X position on the electronic structure. In Table 3, methyl acrylate is given as an example, and H at the α-position is replaced by the following substituents: F, Cl, CN, CH3 and NO2. These molecules were chosen because their Michael reactivity has been previously analyzed by Giraldo et al [43].
The atomic populations of the carbonyl group atoms increase during the addition of an electron to the neutral molecule. In the case of X = H, the carbonyl carbon has larger variations than the carbonyl oxygen (Table 3). These variations correspond to 28.8% of the overall variation. The atomic populations of the carbon-hydrogens at β-position, carbon-hydrogen at α-position, carbons at β- and α-position, and OCH3 respectively vary by 22.5%, 11.1%, 24.6% and 12.9%. These values show that the added electron is mostly located at the carbonyl bond.
Table 3 shows the changes of the atomic populations at Cβ and Ccarbonyl. The population differences at Cβ increase in the order X = NO2 < CH3 < CN < Cl < H < F. At Ccarbonyl the population differences increase in the order X = NO2 < CN < Cl < CH3 < H < F. None of these population differences correlates well with the activation energies of the Cβ attack of methanethiol investigated by Giraldo et al, which increase in the order X = NO2 < CN < H < Cl < CH3 < F. The changes of the atomic populations of the substituent in X position increase in the order F < H < Cl < CH3 < CN < NO2, indicating an increasing electron-withdrawing effect of these substituents. The population differences between Ccarbonyl and Cβ decrease accordingly. This trend shows that Cβ becomes more electrophilic. The values of D and D∩ for these molecules increase in the order F < H < Cl < CH3 < CN < NO2. This correlates reasonably well with the calculated activation energies in ref. [43].
Figure 3 shows that the activation energies are inversely proportional to D. Only the relation with the D descriptor is shown because D and D∩ have the same values for mono(meth)acrylates (Table 2). This suggests that the reactivities of these molecules are correlated with the electron withdrawing effect of the substituent. This can be observed with the NO2 functional group. The lower the activation energy, the larger is the electron withdrawing effect. In consequence, Cβ becomes more electrophilic. In contrast, Giraldo et al did not find any trend between the positive Fukui function of the electron density and the activation energy. They only found for NO2 that it is the most electron-withdrawing substituent, and consequently Cβ has the largest electrophilic character with this substituent [43].
Relation between the activation energy and the descriptor D. The values of D and D∩ are equal for mono(meth)acrylates. Activation energies were taken from ref. [43]. Equation for the adjusted line: y = − 29.376x + 37.242, R.2 = 0.727
3.3 Effects of the substituent in Y position
The diversity of the substituents is broader in Y position than in X position (Scheme 1 and Table 1). We have analyzed acrylates, methacrylates, diacrylates and dimethacrylates with different n-alkyl substituents in Y position to evaluate the correlation with the descriptors proposed in this paper. Substituents with heteroatoms, unsaturated or branched substituents are not included here, because they do not exhibit clear trends.
Figure 4 shows that both descriptors differentiate between acrylates and methacrylates, and diacrylates and dimethacrylates. D and D∩ correlate well with the number of carbon atoms in Y position. Moreover, the values of D are larger in diacrylates and dimethacrylates than in their monosubstituted counterparts. This difference is about 3.4 units. In the case of the monosubstituted molecules, the values of these descriptors are very similar, see Fig. 4 top. Only the relation with the D descriptor is shown because D and D∩ have the same values for mono(meth)acrylates (Table 2). In the same vein, methacrylates have larger values than acrylates by about 0.1 units for both descriptors. These trends show that N(Ω) of the acrolein backbone decreases and is transferred to the n-alkyl group in Y position as the size of the substituent is increased.
Values of D and D∩ versus number of carbon atoms of the alkyl substituent in Y position. A: acrylates (circles) and methacrylates (diamonds). B: diacrylates (circles) and dimethacrylates (diamonds). C: diacrylates (circles) and dimethacrylates (diamonds). Equations for the adjusted lines: a y = 0.008x + 0.412 R2 = 0.731 (methacrylates) y = 0.004x + 0.318, R2 = 0.730 (acrylates); b y = 0.108x + 3.811, R2 = 0.991 (dimethacrylates), y = 0.128x + 3.180, R2 = 0.465 (diacrylates); c y = 0.026x + 0.454, R2 = 0.990 (dimethacrylates) and y = 0.022x + 0.305, R2 = 0.792 (diacrylates). The number on each data point corresponds to the molecule number presented in Table 1
It is important to stress out that the clear separation between different classes of acrylates, and the correlation with the number of carbon atoms in the substituent in Y position, is only found for a subset of the acrylates investigated in this study, namely those with aliphatic and unbranched substituents.
3.4 Hierarchical clustering
As a further test for descriptors D and D∩ (Table 2), a hierarchical cluster analysis was carried out with the set of acrylates shown in Table 1. The best dendrogram was obtained using the Minkowski distance as the similarity function and the grouping methodology of average.
The dendrogram was constructed using the D and D∩ descriptors (Table 2). In the dendrogram, the main observed groups differ because of the D descriptor. But the order within each group is due to D∩. Moreover, in Fig. 5, five main clusters of acrylates can be distinguished: cluster 1 and cluster 2 contain triacrylate and trimethacrylate. Moreover, compounds 13, 15 and 40 contain three acrolein backbones, 13 has a pentaerythritol substituent, and compounds 15 and 40 have a trimethylolpropane substituent (Table 1 and S1). In cluster 3 acrylates with electron-rich substituents in position Y, such as Br or bromobenzyl are found. Cluster 4 includes only monoacrylates and monomethacrylates. Cluster 5 includes diacrylates and dimethacrylates. This shows that our descriptors can classify acrylates based on their chemical nature. Moreover, contiguously grouped molecules indicate that their substituent group withdraws almost the same atomic population from the acrolein backbone. In the same vein, molecules in cluster 3 have substituents that most strongly withdraw atomic population from the acrolein backbone as it can be seen at their D∩ values (Table 2).
The mutagenic compounds (17, 19, 22, 38, 44, 46, 63 and 64) and compounds without reported mutagenicity (56, 57, 58, 59, 60, 61, 62 and 65) are in clusters 3 and 4 (Table S2). In cluster 3 are the compounds with the largest D∩ values (Table 2). This means that the atomic populations of molecules 56 and 57 are localized on the substituents rather than in the acrolein backbone. Moreover, the largest D∩ value for a mutagenic acrylate correspond to compound 38 (Table 1 and 2) which suggests that compounds 56 and 57 may be mutagenic too and this mutagenicity activity should be experimentally evaluated elsewhere.
4 Conclusions
Two descriptors, D and D∩, based on QTAIM atomic populations N(Ω) were proposed and applied to a set of 65 acrylates. These two descriptors D and D∩, based on QTAIM atomic populations, N(Ω), consider the electron-withdrawing effect of the acrolein moiety. The descriptor D is more sensitive to differences in the type of the acrylate than D∩. Applied to a subset of compounds including only aliphatic substituents without heteroatoms the descriptor D differentiates between mono-, di- and tri- acrolein backbones and both descriptors differentiate between acrylates and methacrylates with one or two acrolein backbones.
A cluster analysis using both descriptors shows that the molecules studied herein can be grouped into mono-, di- and tri- acrolein backbones. Mono-acrylates with electron-rich substituents (aromatic groups and halogens) are in a separate group.
Molecules with similar D and D∩ values are characterized by substituent groups that withdraw a similar amount of atomic electron population from the acrolein backbone.
The proposed descriptors describe the atomic population distribution of the mutagenic acrylates and this population is localized on the substituents rather than on the acrolein backbone.
Analyzing the effect of the substituent in the α-carbon (X position) in the methyl acrylate (X: F, Cl, CN, CH3 and NO2) on the changes of their atomic populations, we noticed that these changes increase in the order F < H < Cl < CH3 < CN < NO2, indicating an increasing electron-withdrawing effect of these substituents from the acrolein backbone. In consequence, Cβ becomes more electrophilic. It can be seen that the lower the activation energy, the larger is the electron withdrawing effect of the substituents. Therefore, an electrophilic Cβ favors the Michael reaction [17, 43]. Our results allow us to suggest that 2-(2,4,6-tribromophenoxy)ethyl acrylate and 2,4,6-tribromophenyl acrylate are mutagenic.
The proposed descriptors are useful to understand the atomic property distribution in a set of molecules which share a common backbone. They also may be used in non-supervised machine learning techniques, such as hierarchical clustering to predict the mutagenicity of potential mutagens.
References
Ajekwene KK (2020) Properties and applications of acrylates. In: Serrano-Aroca Á, Deb S (eds) Acrylate polymers for advanced applications. IntechOpen, London
Yu X, Yi B, Wang X (2008) Quantitative structure–property relationships for the reactivity parameters of acrylate monomers. Eur Polym J 44:3997–4001. https://doi.org/10.1016/j.eurpolymj.2008.09.028
Pérez-Garrido A, Morales Helguera A, Giron Rodriguez F, Cordeiro MNDS (2010) QSAR models to predict mutagenicity of acrylates, methacrylates and α, β-unsaturated carbonyl compounds. Dent Mater 26:397–415. https://doi.org/10.1016/j.dental.2009.11.158
Mauri A, Consonni V, Pavan M, Todeschini R (2006) Dragon software: an easy approach to molecular descriptor calculations. Match 56:237–248
Tanış E, Çankaya N, Yalçın S (2019) Synthesis, characterization, computation of global reactivity descriptors and antiproliferative activity of N-(4-nitrophenyl)acrylamide. Russ J Phys Chem B 13:49–61. https://doi.org/10.1134/S1990793119010147
Furuhama A, Aoki Y, Shiraishi H (2012) Development of ecotoxicity QSAR models based on partial charge descriptors for acrylate and related compounds. SAR QSAR Environ Res 23:731–749. https://doi.org/10.1080/1062936X.2012.719542
Ishihara M, Fujisawa S (2009) A structure-activity relationship study on the mechanisms of methacrylate-induced toxicity using NMR chemical shift of β-carbon, RP-HPLC log P and semiempirical molecular descriptor. Dent Mater J 28:113–120. https://doi.org/10.4012/dmj.28.113
Ishihara M, Fujisawa S (2008) Quantum-chemical descriptors for estimating hemolytic activity of aliphatic and aromatic methacrylates. Chemosphere 70:1898–1902. https://doi.org/10.1016/j.chemosphere.2007.07.070
Liu W, Yi P, Tang Z (2006) QSPR models for various properties of polymethacrylates based on quantum chemical descriptors. QSAR Comb Sci 25:936–943. https://doi.org/10.1002/qsar.200510177
Lawson RG, Jurs PC (1990) Cluster analysis of acrylates to guide sampling for toxicity testing. J Chem Inf Comput Sci 30:137–144. https://doi.org/10.1021/ci00066a007
Bader RFW (1991) A quantum theory of molecular structure and its applications. Chem Rev 91:893–928. https://doi.org/10.1021/cr00005a013
Bader RFW (1994) Atoms in molecules: a quantum theory. Clarendon Press, Oxford
Bader RFW (2005) The quantum mechanical basis of conceptual chemistry. Monatshefte Fur Chem - Chem Mon 136:819–854. https://doi.org/10.1007/s00706-005-0307-x
Popelier PLA (2000) Atoms in molecules: an introduction. Prentice Hall, Pearson
Mandado M, González-Moa MJ, Mosquera RA (2007) QTAIMn-center delocalization indices as descriptors of aromaticity in mono and poly heterocycles. J Comput Chem 28:127–136. https://doi.org/10.1002/jcc.20468
Mortelmans K, Zeiger E (2000) The Ames Salmonella/microsome mutagenicity assay. Mutat Res Mol Mech Mutagen 455:29–60. https://doi.org/10.1016/S0027-5107(00)00064-6
Allen TEH, Grayson MN, Goodman JM et al (2018) Using transition state modeling to predict mutagenicity for michael acceptors. J Chem Inf Model 58:1266–1271. https://doi.org/10.1021/acs.jcim.8b00130
National Toxicology Program (NTP) (2019) Ames conclusions dataset. 022–00002–0001–000–007
PubChem Chemical Carcinogenesis Research Information System (CCRIS) [SourceNa - PubChem Substance - NCBI. https://www.ncbi.nlm.nih.gov/pcsubstance/?term=%22Chemical+Carcinogenesis+Research+Information+System+(CCRIS)%22%5BSourceName%5D. Accessed 9 Dec 2022
OECD Work on Co-operating in the Investigation of High Production Volume Chemicals - HPV Database search. https://hpvchemicals.oecd.org/ui/Search.aspx. Accessed 9 Dec 2022
Zeiger E, Anderson B, Haworth S et al (1987) Salmonella mutagenicity tests: III. Results from the testing of 255 chemicals. Environ Mutagen 9:1–60. https://doi.org/10.1002/em.2860090602
Canter DA, Zeiger E, Haworth S et al (1986) Comparative mutagenicity of aliphatic epoxides in Salmonella. Mutat Res Toxicol 172:105–138. https://doi.org/10.1016/0165-1218(86)90069-8
Waegemaekers THJM, Bensink MPM (1984) Non-mutagenicity of 27 aliphatic acrylate esters in the Salmonella-microsome test. Mutat Res Toxicol 137:95–102. https://doi.org/10.1016/0165-1218(84)90097-1
Seifried HE, Seifried RM, Clarke JJ et al (2006) A compilation of two decades of mutagenicity test results with the Ames Salmonella typhimurium and L5178Y mouse lymphoma cell mutation assays. Chem Res Toxicol 19:627–644. https://doi.org/10.1021/tx0503552
Schweikl H, Schmalz G, Rackebrandt K (1998) The mutagenic activity of unpolymerized resin monomers in Salmonella typhimurium and V79 cells. Mutat Res Toxicol Environ Mutagen 415:119–130. https://doi.org/10.1016/S1383-5718(98)00067-9
Geurtsen W (2000) Biocompatibility of resin-modified filling materials. Crit Rev Oral Biol Med 11:333–355. https://doi.org/10.1177/10454411000110030401
Gordon SC, Zimmerman DD, Griffith FD (1991) Acute toxicity, genotoxicity, and dermal carcinogenicity assessment of Isooctyl acrylate. J Toxicol Environ Health 34:297–308. https://doi.org/10.1080/15287399109531569
Zeiger E, Anderson B, Haworth S et al (1988) Salmonella mutagenicity tests: IV. Results from the testing of 300 chemicals. Environ Mol Mutagen 11:1–18. https://doi.org/10.1002/em.2850110602
OECD Work on Co-operating in the Investigation of High Production Volume Chemicals - Chemical Detailed Results - CAS 818–61–1. https://hpvchemicals.oecd.org/UI/handler.axd?id=f5e00c2a-4a2f-4d17-a5c7-5c5853cbdd5c. Accessed 24 Nov 2022
PubChem Chemical Carcinogenesis Research Information System. Cas No. 999-55-3. https://pubchem.ncbi.nlm.nih.gov/substance/363902695. Accessed 24 Nov 2022
OECD Work on Co-operating in the Investigation of High Production Volume Chemicals - Chemical Detailed Results - CAS 7534–94–3. https://hpvchemicals.oecd.org/ui/handler.axd?id=512a6b18-79ca-4eab-93c8-9a7acc602670. Accessed 24 Nov 2022
Pauling L, University C, Press CU (1960) The nature of the chemical bond and the structure of molecules and crystals an introduction to modern structural chemistry. Cornell University Press, Ithaca
Pracht P, Bohle F, Grimme S (2020) Automated exploration of the low-energy chemical space with fast quantum chemical methods. Phys Chem Chem Phys 22:7169–7192. https://doi.org/10.1039/C9CP06869D
Bannwarth C, Ehlert S, Grimme S (2019) GFN2-xTB—an accurate and broadly parametrized self-consistent tight-binding quantum chemical Method with multipole electrostatics and density-dependent dispersion contributions. J Chem Theory Comput 15:1652–1671. https://doi.org/10.1021/acs.jctc.8b01176
Zhao Y, Truhlar DG (2008) The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor Chem Acc 120:215–241. https://doi.org/10.1007/s00214-007-0310-x
Ditchfield R, Hehre WJ, Pople JA (1971) Self-consistent molecular-orbital methods. IX. An extended Gaussian-type basis for molecular-orbital studies of organic molecules. J Chem Phys 54:724–728. https://doi.org/10.1063/1.1674902
Grimme S, Antony J, Ehrlich S, Krieg H (2010) A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J Chem Phys 132:154104. https://doi.org/10.1063/1.3382344
Neese F, Wennmohs F, Becker U, Riplinger C (2020) The ORCA quantum chemistry program package. J Chem Phys 152:224108. https://doi.org/10.1063/5.0004608
Neese F (2022) Software update: The ORCA program system—Version 5.0. WIREs Comput Mol Sci. https://doi.org/10.1002/wcms.1606
Moss GP (1996) Basic terminology of stereochemistry (IUPAC Recommendations 1996). Pure Appl Chem 68:2193–2222. https://doi.org/10.1351/pac199668122193
Keith TA (2019) AIMAll (Version 19.10.12), TK Gristmill Software, Overland Park KS, USA
Müller P (1994) Glossary of terms used in physical organic chemistry (IUPAC Recommendations 1994). Pure Appl Chem 66:1077–1184. https://doi.org/10.1351/pac199466051077
Giraldo C, Gómez S, Weinhold F, Restrepo A (2016) Insight into the Mechanism of the Michael reaction. ChemPhysChem 17:2022–2034. https://doi.org/10.1002/cphc.201600166
Müllner D (2013) Fastcluster: Fast Hierarchical, Agglomerative Clustering Routines for R and Python. J Stat Softw. https://doi.org/10.18637/jss.v053.i09
Kaufman L, Rousseeuw PJ (1990) Finding groups in data. Wiley, Hoboken, NJ, USA
Lance GN, Williams WT (1967) A general theory of classificatory sorting strategies: 1. Hierarchical Syst Comput J 9:373–380. https://doi.org/10.1093/comjnl/9.4.373
R Core Team (2021) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Galili T (2015) dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 31:3718–3720. https://doi.org/10.1093/bioinformatics/btv428
Saracli S, Doğan N, Doğan İ (2013) Comparison of hierarchical cluster analysis methods by cophenetic correlation. J Inequalities Appl 2013:203. https://doi.org/10.1186/1029-242X-2013-203
Acknowledgements
This work was supported by project C-2021–01 from Universidad Industrial de Santander.
Funding
Open Access funding provided by Colombia Consortium.
Author information
Authors and Affiliations
Contributions
D.A.R.D. wrote the main manuscript text and performed the calculations, A.J.E. contributed with the information deposited in the acrylates dataset. D.A.R.D., M.D. and M.C.D. analyzed the data and reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rincón, D.A., Escorcia, A.J., Doerr, M. et al. QTAIM based descriptors for the classification of acrylates. Theor Chem Acc 142, 131 (2023). https://doi.org/10.1007/s00214-023-03034-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00214-023-03034-4