Abstract
An in depth investigation at the genomic level is needed to identify early human-relevant cardiotoxicity biomarkers that are induced by drugs and environmental toxicants. The main objective of this study was to investigate the role of microRNAs (miRNAs) as cardiotoxicity biomarkers using human-induced pluripotent stem cell (hiPSC)-derived cardiomyocytes (CMs) that were exposed to doxorubicin (DOX) as a “gold standard” cardiotoxicant. hiPSC-CMs were exposed to 156 nM DOX for 2 days or for 6 days of repeated exposure, followed by drug washout and incubation in drug-free culture medium up to day 14 after the onset of exposure. The induced miRNAs were profiled using miRNA microarrays, and the analysis of the data was performed using the miRWalk 2.0 and DAVID bioinformatics tools. DOX induced early deregulation of 14 miRNAs (10 up-regulated and 4 down-regulated) and persistent up-regulation of 5 miRNAs during drug washout. Computational miRNA gene target predictions suggested that several DOX-responsive miRNAs might regulate the mRNA expression of genes involved in cardiac contractile function. The hiPSC-CMs exposed to DOX in a range from 39 to 156 nM did not show a significant release of the cytotoxicity marker lactate dehydrogenase (LDH) compared to controls. Quantitative real-time PCR analyses confirmed the early deregulation of miR-187-3p, miR-182-5p, miR-486-3p, miR-486-5p, miR-34a-3p, miR-4423-3p, miR-34c-3p, miR-34c-5p and miR-1303, and also the prolonged up-regulation of miR-182-5p, miR-4423-3p and miR-34c-5p. Thus, we identified and validated miRNAs showing differential DOX-responsive expression before the occurrence of cytotoxicity markers such as LDH, and these miRNAs also demonstrated the significant involvement in heart failure in patients and animal models. These results suggest that the DOX-induced deregulated miRNAs in human CMs may be used as early sensitive cardiotoxicity biomarkers for screening potential drugs and environmental cardiotoxicants with a similar mechanism of action.
Similar content being viewed by others
Introduction
Irreversible cardiac injury is a most serious side effect associated with many known anti-cancer drugs and other therapeutic drugs, putting patients’ lives at risk. Furthermore, over the last two decades, the pharmaceutical industry has been struggling with the costly withdrawals of new drug candidates in late-stage clinical trials. Thus, one of the major tasks of pharmaceutical companies is to predict the cardiotoxic side effects of new drug candidates early in non-clinical phases of drug development. Moreover, it has been recognized since long time that environmental toxicants and pollutants may induce cardiotoxicity in animals and humans (Simkhovich et al. 2009). Several preclinical models have been developed to understand doxorubicin (DOX)-induced early and chronic cardiotoxicity (AbdelAleem et al. 1997; Banco et al. 2011; Desai et al. 2013; Hrelia et al. 2002; Jaenke 1974; Tokarska-Schlattner et al. 2005). Importantly, all of these established models are non-human in origin. Due to an inter-species variation in physiology, assays using these established models cannot reliably predict the cardiotoxic effects of new drugs in humans. However, current studies have demonstrated that in vitro use of human embryonic stem cell (hESC)- or human-induced pluripotent stem cell (hiPSC)-derived cardiomyocytes can be beneficial for preclinical safety assessment.
Currently applied diagnostic techniques [ecocardiographic left ventricular ejection fraction (LVEF), radionuclide angiography and endomyocardial biopsy] for assessing drug-induced cardiac injury have many shortcomings, including sensitivity, invasiveness and high cost. In preclinical and clinical phase studies, troponins (cTnT and cTnI) have been used as highly sensitive plasma biomarkers for detecting DOX-induced cardiac damage (Herman et al. 1998, 1999; Lipshultz et al. 2012; O’Brien 2006; O’Brien et al. 2006; Shahzadi et al. 2014). However, troponins and many other cardiac biomarkers show increased plasma levels only after heart tissue damage and also have a limited half-life (Tonomura et al. 2009; Walker 2006). Considering the disadvantages of current biomarkers, there is an urgent need to identify stable, highly sensitive and noninvasive genomic biomarkers to predict early events of drug-induced cardiac damage.
Among anthracyclines, DOX is a highly effective anti-cancer drug prescribed for the treatment of a variety of cancer types, including solid tumours and haematologic malignancies in both adults and children. Despite its beneficial therapeutic effects, the long-term clinical use of DOX is limited, due to its cumulative dose-dependent cardiotoxicity (Singal and Iliskovic 1998). A recent meta-analysis demonstrates that anthracycline treatment increased the risk of cardiac death by 4.94-fold compared to non-anthracycline treatments (Smith et al. 2010). The acute adverse effects of DOX occur within or after 2–3 days of administration and are manifested by cardiac arrhythmias and acute heart failure. The chronic side effects of DOX are largely dose-dependent. A patient may develop dilated cardiomyopathy shortly after DOX treatment termination, or dilated cardiomyopathy may occur even 10–15 years after the termination of chemotherapy. Acute and chronic DOX administration may lead to cardiac dysfunction, cardiomyopathy and ultimately to heart failure and death (Chatterjee et al. 2010; Wallace 2003; Yeh et al. 2004). DOX is a well-characterized cardiotoxicant of the anthracycline family, but the exact mechanisms of DOX-induced cardiotoxicity are not fully understood. Nevertheless, DOX can be applied as a substance to identify human-relevant genomic biomarkers for potential cardiotoxic drugs and environmental factors that induce cardiotoxicity (Chaudhari et al. 2015).
Small non-coding RNAs that are approximately 22 nucleotides long are known as microRNAs (miRNAs) that regulate gene expression by targeting messenger RNAs (mRNAs) by binding to complementary regions of transcripts to inhibit their translation and/or promote mRNA degradation (Ambros 2004; Bartel 2004). In particular, miRNAs are significantly involved in heart development, physiology and pathogenesis (Latronico and Condorelli 2009; Rao et al. 2009; Small et al. 2010a). These functions make miRNAs an attractive target for identifying sensitive biomarkers of cardiovascular diseases (Bernardo et al. 2012; D’Alessandra et al. 2010; Matsumoto et al. 2013).
Here, for the first time, we identify set of miRNAs as toxicity signatures of DOX in hiPSC-cardiomyocytes (CMs). In the present study, we aimed to identify early and prolonged differential expression of miRNAs in response to DOX exposure in hiPSC-CMs by profiling global miRNA levels. Our miRNA microarray results identified 14 early DOX-responsive miRNAs (10 up-regulated and 4 down-regulated), and five miRNAs that are persistently up-regulated during drug washout. DOX-deregulated miRNAs and their in silico predicted gene targets are associated with intact cardiac function and cardiac diseases. Our results demonstrate that early and/or prolonged deregulated miRNAs are promising broad biomarkers of cardiotoxicity that also provide information regarding the early events of cardiac damage.
Materials and methods
Cardiomyocyte cell culture
All experiments were performed using iCell Cardiomyocytes® (Cellular Dynamics International, Madison, WI, USA) derived from hiPSCs. A 98 % pure population of cardiomyocytes was supplied in a cryopreserved single-cell suspension. The cardiomyocytes were a mixture of electrically active atrial-, nodal- and ventricle-like myocytes. Cryopreserved cardiomyocytes were thawed and plated on fibronectin-coated (5 µg/cm2, 2 h at 37 °C) (Sigma-Aldrich, Steinheim, Germany) 6-well plates and 96-well plates using iCell Cardiomyocytes Plating Medium (iCell-PM) (Cellular Dynamics International, Madison, WI, USA). Two days post-plating, cells were cultured in iCell Cardiomyocytes Maintenance Medium (iCell-MM) (Cellular Dynamics International, Madison, WI, USA), with fresh media changed every 2 days.
Test compound
A 10 mM stock solution (in DMSO) of DOX was purchased from Selleck chemicals. The stock solution was dispensed into small volume aliquots and stored at −80 °C. The drug dilutions were performed in iCell-MM at room temperature prior to each drug exposure.
DOX exposure and cell sample collection for microarray studies
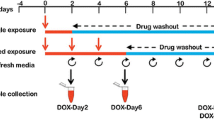
A schematic representation of the experimental design is shown in Fig. 1. Briefly, the iCell cardiomyocytes were cultured in a 6-well plate at a density of 0.4 × 106 cells per well. Four days post-plating, the synchronously beating cardiomyocytes were incubated with DOX at 156 nM for a 2-day exposure period (DOX-Day2) or three consecutive exposure periods of DOX for 6 days (DOX-Day6) (DOX supplemental media was refreshed every 48 h). Following DOX exposure for a 2-day washout (DOX-Day2WO) or a 6-day washout (DOX-Day6WO), cardiomyocytes were further cultivated in DOX-free iCell-MM until day 14 (from the start of the drug exposure). Non-treated control cells were cultured for 6 days in iCell-MM with DMSO as the solvent. The culture medium was refreshed every 2 days. The control, DOX-exposed and drug washout cell samples were harvested on day 2, 6 and 14, respectively, using QIAzol lysis reagent (Qiagen, Hilden, Germany). Total RNA, including non-coding miRNAs, were extracted from harvested cell samples and used for miRNA microarray hybridization and real-time PCR studies.
Experimental design for repeated drug exposure to hiPSC-CMs. Synchronously beating cardiomyocytes were given a single DOX exposure for 2 days or repeated DOX exposure for 6 days (the DOX-supplemented culture media was refreshed every 2 days). Following single and repeated drug exposure, the cells were cultured in drug-free culture media until day 14 (after the onset of drug exposure). During the drug washout, the culture medium was refreshed every 2 days. For microarray studies, cell samples were harvested on day 2, 6 and 14. This figure was reproduced from our previous report (Chaudhari et al. 2015)
Total RNA extraction
The total RNA, including non-coding miRNAs, were isolated from control, DOX-exposed and drug washout cardiomyocytes using the miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Concentration and purity of the isolated RNA was evaluated using a Nanodrop ND-1000 spectrophotometer (ND-1000, Thermo Fisher, Langenselbold, Germany). RNA integrity was confirmed using the Experion™ automated electrophoresis system (Bio-Rad, Munich, Germany).
miRNA microarray labelling and hybridization
The FlashTag Biotin HSR RNA labelling Kit (Affymetrix, High Wycombe, United Kingdom) was used to label 500 ng of total RNA. The biotin labelled samples were hybridized to GeneChip miRNA 3.0 arrays (Affymetrix, High Wycombe, United Kingdom) using a hybridization cocktail. Microarray hybridization was performed in an Affymetrix GeneChip Hybridization Oven-645 for 16 h at 48 °C and 60 rpm. Hybridized arrays were washed and stained using the GeneChip HWS Kit (Affymetrix, High Wycombe, United Kingdom) and the fluidics station protocol (FS450_0002) on an Affymetrix GeneChip Fluidics Station-450. The stained arrays were scanned with an Affymetrix GeneChip Scanner-3000-7G, and quality of the scanned arrays was evaluated using Affymetrix GCOS software. The generated raw data files were used for data analysis.
miRNA microarray data analysis
The miRNA expression data procured from the GeneChip® miRNA 3.0 arrays were quantile normalized using the RMA approach with the Partek® Genomics Suite® software, version 6.6 (Copyright Partek Inc., St. Louis, MO, USA). The probe sets for 5787 miRNAs specific to Homo sapiens were selected further for statistical calculation. The differential expressions between groups were analysed for DOX-Day2 versus Control-Day2, DOX-Day6 versus Control-Day6, and DOX-Day2WO and DOX-Day6WO versus Control-Day14. Statistical calculations to determine significant genes were executed with the linear model implementation of the R Limma package followed by a Benjamini-Hochberg multiple test correction (1 % FDR). The miRNAs with a minimum fold change 1.8 and p value <0.05 were selected for further data analysis.
Prediction of miRNA-gene targets
The gene target prediction of perturbed miRNAs was performed using the miRWalk 2.0 database (Dweep et al. 2011). Unlike currently available miRNA-gene target predictive tools, miRWalk 2.0 can identify putative miRNA binding sites not only in the 3′-UTR region but also in the promoter, the 5′-UTR and the CDS (amino acid coding sequence) regions of a gene. The miRWalk database is updated routinely and also provides information on validated miRNA binding sites in human genes. The predicted gene targets of the miRNAs were systematically compared and verified with our previously reported DOX transcriptomic data that contains differentially expressed genes (fold change of 2.0, FDR® p value <0.05) for DOX-Day2, DOX-Day6, DOX-Day2WO and DOX-Day6WO groups (Chaudhari et al. 2015) (Fig. 2a). The predicted gene targets of the up-regulated miRNAs were verified with commonly down-regulated genes among the DOX-Day2 and DOX-Day6 groups, while the predicted gene targets of down-regulated miRNAs were confirmed by comparison with the commonly up-regulated genes between the DOX-Day2 and DOX-Day6 groups (Fig. 2b). Similarly, the predicted gene targets of the persistently up-regulated miRNAs were verified with prolonged down-regulated genes (Fig. 2c). Verified gene targets from the transcriptome data were used for Gene ontology (GO) analysis. The GO enrichment and KEGG pathway analyses were performed using the online Database for Annotation, Visualization and Integrated Discovery (DAVID) programme (Dennis et al. 2003).
a Flow chart of the microarray data analysis used in this work. Differentially expressed miRNAs and their putative gene targets were verified with gene expression (mRNA) data subsequently the verified gene targets were used for the GO analysis. b, c Overlapping genes from the transcriptomic data matched with the miRNA gene targets predicted using miRWalk 2.0. Venn diagrams show that the predicted gene targets of the up-regulated miRNAs matched with the overlapping down-regulated genes and vice versa. Confirmed gene targets used for GO analysis
Quantitative real-time PCR (qPCR)
Using 500 ng of total RNA, cDNA synthesis was performed with the qScript™ microRNA cDNA Synthesis Kit (Quanta Biosciences, Gaithersburg, USA) following the manufacturer’s instructions. The cDNA was diluted fivefold with nuclease-free water, and 1 µl was used as a template for qPCR. The amplification of miRNA was performed using the PerfeCTa® microRNA assay primer, the PerfeCTa® Universal PCR primer and the PerfeCTa® SYBR® Green SuperMix, Low ROX™ Kit (Quanta Biosciences, Gaithersburg, USA) as per the manufacturer’s instructions. qPCR was carried out on an Applied Biosystems 7500 FAST Real-Time PCR System. Relative miRNA levels were calculated using the ΔΔC t method, and RNU6 was used as the miRNA PCR control.
LDH leakage assay
Lactate dehydrogenase (LDH) is a widely used marker to evaluate the presence of damage and toxicity in cells and tissues. LDH is a cytosolic enzyme that is not normally discharged outside of the cell, but upon plasma membrane damage of cells, LDH is released into the cell culture medium or blood. Measurement of extracellular LDH release into the culture medium can be used to assay cellular toxicity. The iCell cardiomyocytes were seeded on a fibronectin-coated (5 µg/cm2, 2 h at 37 °C) 96-well plate at a cell density of 20 × 103 per well. On day 4 post-seeding, cardiomyocytes were exposed to DOX at different concentrations. After 48 h of exposure, extracellular LDH activity was measured using the Thermo Scientific™ Pierce™ LDH Cytotoxicity Assay Kit according to the manufacturer’s instructions. Absorbance was measured at 490 nm using the Softmax Pro M5e 96-well plate reader (Molecular Devices, Sunnyvale, CA, USA). LDH activity was expressed as fold change versus control samples.
Results
Repeated exposure to DOX-induced cytotoxicity
Similar to our previous report (Chaudhari et al. 2015), repeated exposure to DOX induced significant cytotoxicity at day 6 (DOX-Day6) compared to Control-Day6 cardiomyocytes (Fig. 3a). This result is indicative of a decrease in cell viability and ultimately cardiac cell loss. During drug washout, especially in repeatedly exposed cardiomyocytes (DOX-Day6WO), a slight, but not significant, decrease in cell viability was observed compared to the DOX-Day6 group. A single exposure of DOX did not induce a significant level of cytotoxicity in DOX-Day2 and DOX-Day2WO groups compared to the Control-Day2 and Control-Day14 cardiomyocytes, respectively. Additionally, no significant increase in LDH leakage was observed after 2 days of exposure to DOX at a concentration of 156 nM compared to controls (Fig. 3b). However, exposure to DOX at a concentration above 156 nM resulted in progressive increase in LDH release, which indicated the presence of cell membrane damage and cytotoxicity. These findings demonstrate that a 2-day exposure of DOX at 156 nM to cardiomyocytes could be used to identify early changes in miRNA expression levels under less-adverse cytotoxic conditions.
a Effect of DOX (156 nM) single and repeated exposure on cell viability. Cell images show that compared to controls, hiPSC-CM viability neither changed during single DOX exposure (DOX-Day2) nor during drug washout in DOX-Day2WO group. However, compared to controls, repeated exposure to DOX induced significant cytotoxicity and resulted in decreased cell viability in the DOX-Day6 group. During drug washout, slightly more cell death was observed in the DOX-Day6WO group compared to the DOX-Day6 group. Scale bar indicates 50 µm. B, DOX-induced cytotoxicity was assessed by LDH leakage assay. The hiPSC-CMs were incubated with DOX for 48 h at a concentration range of 0.039–1.25 µM. The enzymatic activity was analysed for the cellular release of LDH into the culture media. The bar graph shows mean values of fold change compared to controls, and the error bars indicate the standard deviation (n = 4). ***p value <0.001 versus control group
miRNA profiling identified early DOX-responsive and persistently deregulated miRNAs
The miRNA microarray data analysis resulted in the identification of several differentially expressed miRNAs in the DOX-treated and washout groups (Fig. 4a). A single exposure (DOX-Day2) and a repeated exposure (DOX-Day6) to DOX perturbed, in total, 21 (15 up-regulated and seven down-regulated) and 79 (37 up-regulated and 42 down-regulated) miRNAs, respectively. The five up-regulated miRNAs in the DOX-Day2WO group and the 26 miRNAs (15 up-regulated and 11 down-regulated) in the DOX-Day6WO group showed prolonged differential expression during drug washout. Differentially expressed miRNAs under different experimental conditions are provided in Supplemental Table S1. A Venn diagram analysis was performed for DOX-exposed and drug washout groups to identify commonly deregulated miRNAs. The analysis identified 14 (10 up-regulated and four down-regulated) commonly deregulated miRNAs between the DOX-Day2 and DOX-Day6 groups (Fig. 4b, c). These 14 deregulated miRNAs are indicative of an early response to DOX, and their fold change values are provided in Table 1. Among the DOX-exposed and drug washout groups, five miRNAs showed persistent up-regulation (Fig. 4d, and their fold change values are provided in Table 1), while no commonly down-regulated miRNAs were observed (Fig. 4e).
a Number of differentially expressed miRNAs in different experimental conditions. Three independent experiments were performed for miRNA microarray studies. Comparisons with respective control groups were performed to identify the DOX-induced differentially expressed miRNAs (with at least a fold change 1.8 and a p value <0.05 vs. control). The number of up- and down-regulated miRNAs is presented in the bar graph. The miRNAs, those did not deregulate during DOX exposure but showed deregulations during drug washout were omitted from washout groups before the data analysis. b, c Venn diagrams representing the number of commonly up- and down-regulated miRNAs among the DOX-Day2 and DOX-Day6 groups, respectively. d, e Venn diagrams illustrating number of overlapping up- and down-regulated miRNAs among the DOX-exposed and washout groups, respectively
Verified miRNA-gene targets enriched in cardiac and stress-associated GOs
Verified gene targets of each miRNA in Table 1 were first identified using the miRWalk 2.0 bioinformatics tool. The putative gene targets of the up-regulated miRNAs were matched with the identified DOX-induced differentially down-regulated genes at day 2, 6 and 14 as previously described (Chaudhari et al. 2015) (see Supplemental Table S2). Common confirmed genes were analysed for the enrichment of GO and KEGG pathways using the DAVID functional enrichment programme. This analysis revealed early DOX-affected cardiac processes, pathways and general toxic responses (Table 2). The down-regulated genes were mainly enriched for GO terms such as the sarcomere, muscle contraction and ion channel activity, and KEGG pathways such as cardiac muscle contraction, HCM, DCM and the MAPK signalling pathway. The putative target genes of the down-regulated miRNAs were matched with the up-regulated genes. Analysis of the common verified genes revealed enrichment for GO terms such as apoptosis, oxidative stress, inflammatory responses and the extracellular region (Table 2). Other GOs are provided in Supplemental Table S3. Verified gene targets of miR-187-3p, miR-486-5p, miR-34a, miR-212-3p, miR-34c-3p, miR-675-5p and miR-3911 were not enriched for GO terms related to cardiac and general toxicity responses. Similarly, verified gene targets of persistently up-regulated miRNAs were not enriched for cardiac-related GOs.
Validation of deregulated miRNA expression using qPCR
The 14 early DOX-responsive miRNAs and the five persistently up-regulated miRNAs were validated using qPCR (for primers sequences see supplemental Table S4). The qPCR data analysis confirmed deregulation of nine miRNAs (8 up and 1 down) in both DOX-Day2 and DOX-Day6 groups (Fig. 5a, b), and also confirmed the prolonged up-regulation of three miRNAs in the washout groups at day 14 (Fig. 5c).
Quantitative real-time PCR (qPCR) validation of miRNA microarray data. The qPCR confirmed significant a up-regulation of eight miRNAs, b down-regulation of miR-1303 in both the DOX-Day2 and DOX-Day6 groups, and c prolonged up-regulation of miR-182-5p, miR-4423-3p and miR-34c-5p in the drug washout groups at day 14. The bar graph represents the fold regulation of miRNAs as the mean ± standard deviation (n = 3, p value <0.05 vs. control)
Discussion
High throughput miRNA profiling is progressively becoming an important screening tool to identify miRNAs that are responsive to drugs and environmental factors. Using this approach, in the present study, we identified early and prolonged DOX deregulated miRNAs that can be used to detect cardiotoxicants. Myocardial apoptosis is one of the main causes of progressive heart failure during DOX treatment. In the present study, repeated exposure to 156 nM DOX-induced substantial cytotoxicity in the DOX-Day6 group, which resulted in cell death and loss of cardiomyocytes. In addition, we also found that compared to controls and a single-DOX exposure group, hiPSC-CMs after repeated exposure to DOX showed arrhythmic beating (Chaudhari et al. 2015). Previous reports have shown that DOX induces myocardial apoptosis in animal models (Fisher et al. 2005; Sharov et al. 1996; Ueno et al. 2006). Increasing evidence suggests that apoptosis is crucially involved in the loss of cardiomyocytes in failing human hearts (Olivetti et al. 1997), myocarditis (Kawano et al. 1994) and myocardial infarction (Saraste et al. 1997). Ageing is also associated with a gradual decrease in cardiac performance and a loss of cardiomyocytes through apoptosis (Kwak 2013).
In this study, a miRNA microarray analysis revealed 14 early DOX-responsive miRNAs. Of these 14 miRNAs, nine returned to basal levels during drug washout, whereas the other five miRNAs showed persistent up-regulation during drug washout. These results suggest that the effect of DOX on the expression of the majority of miRNAs in cardiomyocytes is reversible if cardiomyocytes are only acutely exposed to DOX.
Validation of miRNA microarray data using qPCR confirmed deregulation of miR-187-3p, miR-182-5p, miR-486-3p, miR-486-5p, miR-34a-3p, miR-4423-3p, miR-34c-3p, miR-34c-5p and miR-1303 in both DOX-Day2 and DOX-Day6 groups. qPCR also confirmed the prolonged up-regulation of miR-182-5p, miR-4423-3p and miR-34c-5p in DOX-Day2WO and DOX-Day6WO groups. These identified deregulated miRNAs showed an early response to DOX that occurred before the induction of general cytotoxic biomarkers such as LDH. A 156 nM DOX treatment for 24 and 48 h did not induce a significant release of cardiac troponin T (cTnT) from hESC-derived cardiomyocytes in culture medium (Andersson et al. 2010; Holmgren et al. 2015). These observations indicate that miRNAs appear to be more sensitive toxicity signatures compared to LDH and other biomarkers such as cardiac troponin in DOX-exposed hiPSC-CMs. qPCR validated the use of the identified miRNAs in further investigations.
The miR-34 family members are involved in cardiac ageing, cardiac diseases and in cardiac apoptotic events. Recently, significant up-regulation of miR-34a has been reported in the aged hearts of mice (Boon et al. 2013), after myocardial infarction in mice (Lin et al. 2010) and after human dilated cardiomyopathy (Elzenaar et al. 2013). This up-regulation of miR-34a is associated with a loss of cardiomyocytes via apoptosis in both myocardial infarction patients and rats (Fan et al. 2013). Higher plasma levels of miR-34a may be a predictive indicator of heart failure after acute myocardial infarction in patients (Matsumoto et al. 2013) and may be a potential biomarker for coronary artery disease (Han et al. 2015). DOX-induced up-regulation of miR-34a has been reported recently in a mouse model (Desai et al. 2014). In agreement with these observations, our results show that DOX-induced up-regulation of miR-34a-3p expression can be correlated with increased apoptosis and decreased cell viability in repeatedly exposed cardiomyocytes (DOX-Day6). These results suggest a potential role of miR-34a-3p in early cytotoxic events in hiPSC-CMs.
Similarly, miR-34b and miR-34c (miR-34 family members) are up-regulated during rat cardiac hypertrophy (Feng et al. 2014), in the aged hearts of mice (Boon et al. 2013), in mouse hearts with myocardial infarction (Bernardo et al. 2012) and in diabetic ischaemic heart failure patients (Greco et al. 2012). DOX-induced up-regulation of miR-34b and miR-34c was reported in a mouse model (Desai et al. 2014) and in rat hearts (Vacchi-Suzzi et al. 2012). Consistent with these observations, our data demonstrated early up-regulation of miR-34c-3p and miR-34c-5p during DOX exposure. Furthermore, miR-34c-5p showed persistent up-regulation after the drug washout. Enriched GOs of verified gene targets of miR-34c-5p included those such as the sarcomere and muscle contraction and verified down-regulated gene targets including ion channels such as KCNN2, SCN5A, CACNA2D2 and KCNK3. As previously shown, these findings can be linked to arrhythmic beating in cardiomyocytes during repeated exposure to DOX (Chaudhari et al. 2015). These observations suggest that up-regulation of miR-34c-3p and miR-34c-5p may be early indicators of cardiac dysfunction, the development of cardiac pathologies and future heart failure.
A higher expression level of miR-486-3p has been found in cats with hypertrophic cardiomyopathy (Weber et al. 2015) and in humans with DCM and HCM (Leptidis et al. 2013). Consistent with these reports, our present study shows an up-regulation of miR-486-3p in DOX-exposed cardiomyocytes. Additionally, the enriched GOs of the verified gene targets of miR-486-3p included those related to cardiac contractile function and ion channel activity. Also, the enriched KEGG pathways included HCM and DCM. Early up-regulation of miR-486-3p could lead to impaired cardiac function, and possibly to cardiomyopathies in hiPSC-CMs.
miR-486 is one of the highly enriched miRNA species in human and mouse hearts (Alexander et al. 2014). Overexpression of miR-486 reduces PTEN levels and activates PI3K/AKT signalling, which eventually leads to cardiac hypertrophy (Small et al. 2010b). Increased expression of miR-486-5p has been found in the plasma of acute myocardial infarction patients (Hsu et al. 2014) and also in cats with hypertrophic cardiomyopathy (Weber et al. 2015). Our findings showing a DOX-induced early up-regulation of miR-486-5p suggest that elevation of miR-486-5p may be considered as an early event in the development of cardiovascular diseases.
Among early up-regulated miRNAs, miR-187-3p showed the highest up-regulation in our miRNA microarray data analysis. Until now, no reports of miR-187-3p in heart failure and DOX-mediated cardiotoxicity have been observed. For the first time, we demonstrated that DOX caused a higher expression of miR-187-3p in human cardiomyocytes. Notably, miR-187-3p overexpressing T-lymphoma cells show resistance to DNA damaging agents such as DOX, cisplatin and cyclophosphamide (Yan et al. 2014). Thus, we may assume that miR-187-3p overexpression in cardiomyocytes is the result of DOX-induced DNA damage. However, more studies are needed to unveil the role of miR-187-3p in both cardiotoxicity and the development of heart failure.
An elevated level of miR-182 was found in patients with coronary disease (Taurino et al. 2010), dilated cardiomyopathy and chronic congestive heart failure (Zhu et al. 2013). DOX-induced DNA damage is an early event of lethal cardiac injury and cell death (L’Ecuyer et al. 2006). Moreover, increased levels of miR-182 impede the DNA repair process and increase genomic instability in cancer cell lines (Krishnan et al. 2013; Moskwa et al. 2011; Yao and Ventura 2011). Additionally, increased expression of miR-182 in melanoma cells after DOX treatment has been reported (Yan et al. 2012). Similarly, in our study, DOX induced the up-regulation of miR-182-5p in hiPSC-CMs, while verified gene targets enriched KEGG pathways such as cardiac muscle contraction. These observations suggest that miR-182-5p may be an initial indicator of cardiac injury and possibly DNA damage. The prolonged up-regulation of this miRNA may explain the mechanism of DOX-induced cardiac diseases and heart failure.
In the present study, we found that DOX induced an early and prolonged up-regulation of miR-4423-3p in hiPSC-CMs. Verified gene targets of miR-4423-3p were significantly enriched in both cardiac-specific GOs and KEGG pathways such as cardiac muscle contraction. Further investigation is needed to understand role of this miRNA in early cardiac damage, and more generally, in the progressive development of cardiac diseases. Our study also showed down-regulation of miRNAs particularly miR-1303 and miR-4298 after DOX exposure. In addition GO enrichment analysis showed that verified gene targets of miR-1303 and miR-4298 significantly enriched GOs associated with stress and apoptosis. However, role of these two miRNAs in development of cardiac pathogenesis or heart failure or ageing needs an in-depth investigation.
As we previously described (Chaudhari et al. 2015), a single exposure of 156 nM DOX compromised the beating heart rate without inducing cytotoxicity. In addition, a 156 nM concentration of DOX is within the therapeutic range of DOX. Repeated exposure to DOX (156 nM) induced substantial cytotoxicity in hiPSC-CMs. The nine validated miRNAs were significantly deregulated with a single exposure to DOX before the indication of the cytotoxicity marker LDH. In the present study, an early up-regulation of miR-34 family members was identified, which indicated that apoptosis could be a very early molecular event that induces cardiac cell loss in hiPSC-CMs after repeated exposure to DOX. Other early up-regulated miRNAs suggest a possible risk of cardiovascular diseases, such as cardiac hypertrophy and future heart failure. Prolonged up-regulation of three miRNAs suggests a risk of cardiac pathophysiology during the long-term recovery phase of DOX-exposed hiPSC-CMs. The DOX-induced miRNA deregulation in hiPSC-CMs is an indicative of complex molecular changes related to apoptosis, ageing, senescence, hypertrophy and other cardiac diseases, which ultimately progress towards cardiac dysfunction and heart failure. The early-deregulated miRNAs identified in this study might be beneficial for the identification and development of early genomic biomarkers of cardiotoxicity in humans and may also be useful for evaluating the risk of cardiac damage caused by potential therapeutic drugs and/or environmental factors with similar mechanism of action.
References
Abdel Aleem S, El Merzabani MM, Sayed Ahmed M, Taylor DA, Lowe JE (1997) Acute and chronic effects of adriamycin on fatty acid oxidation in isolated cardiac myocytes. J Mol Cell Cardiol 29:789–797
Alexander MS, Casar JC, Motohashi N, Vieira NM, Eisenberg I, Marshall JL, Gasperini MJ, Lek A, Myers JA, Estrella EA, Kang PB, Shapiro F, Rahimov F, Kawahara G, Widrick JJ, Kunkel LM (2014) MicroRNA-486 dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy-associated symptoms. J Clin Investig 124:2651–2667
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355
Andersson H, Steel D, Asp J, Dahlenborg K, Jonsson M, Jeppsson A, Lindahl A, Kagedal B, Sartipy P, Mandenius CF (2010) Assaying cardiac biomarkers for toxicity testing using biosensing and cardiomyocytes derived from human embryonic stem cells. J Biotechnol 150:175–181
Banco B, Grieco V, Servida F, Giudice C (2011) Sudden death in a dog after doxorubicin chemotherapy. Vet Pathol 48:1035–1037
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Bernardo BC, Gao XM, Winbanks CE, Boey EJH, Tham YK, Kiriazis H, Gregorevic P, Obad S, Kauppinen S, Du XJ, Lin RCY, McMullen JR (2012) Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci USA 109:17615–17620
Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, Horrevoets AJG, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, Muller OJ, Potente M, Zeiher AM, Hermeking H, Dimmeler S (2013) MicroRNA-34a regulates cardiac ageing and function. Nature 495:107–110
Chatterjee K, Zhang J, Honbo N, Karliner JS (2010) Doxorubicin cardiomyopathy. Cardiology 115:155–162
Chaudhari U, Nemade H, Wagh V, Gaspar JA, Ellis JK, Srinivasan SP, Spitkovski D, Nguemo F, Louisse J, Bremer S, Hescheler J, Keun HC, Hengstler JG, Sachinidis A (2015) Identification of genomic biomarkers for anthracycline-induced cardiotoxicity in human iPSC-derived cardiomyocytes: an in vitro repeated exposure toxicity approach for safety assessment. Arch Toxicol. doi:10.1007/s00204-015-1623-5
D’Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, Rubino M, Carena MC, Spazzafumo L, De Simone M, Micheli B, Biglioli P, Achilli F, Martelli F, Maggiolini S, Marenzi G, Pompilio G, Capogrossi MC (2010) Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J 31:2765–2773
Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4(5):P3
Desai VG, Herman EH, Moland CL, Branham WS, Lewis SM, Davis KJ, George NI, Lee T, Kerr S, Fuscoe JC (2013) Development of doxorubicin-induced chronic cardiotoxicity in the B6C3F1 mouse model. Toxicol Appl Pharmacol 266:109–121
Desai VG, Kwekel JC, Vijay V, Moland CL, Herman EH, Lee T, Han T, Lewis SM, Davis KJ, Muskhelishvili L, Kerr S, Fuscoe JC (2014) Early biomarkers of doxorubicin-induced heart injury in a mouse model. Toxicol Appl Pharmacol 281:221–229
Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk—database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44:839–847
Elzenaar I, Pinto YM, van Oort RJ (2013) MicroRNAs in heart failure: new targets in disease management. Clin Pharmacol Ther 94:480–489
Fan F, Sun AJ, Zhao HT, Liu XW, Zhang WB, Jin XT, Wang C, Ma X, Shen C, Zou YZ, Hu K, Ge JB (2013) MicroRNA-34a promotes cardiomyocyte apoptosis post myocardial infarction through down-regulating aldehyde dehydrogenase 2. Curr Pharm Des 19:4865–4873
Feng HJ, Ouyang W, Liu JH, Sun YG, Hu R, Huang LH, Xian JL, Jing CF, Zhou MJ (2014) Global microRNA profiles and signaling pathways in the development of cardiac hypertrophy. Braz J Med Biol Res 47:361–368
Fisher PW, Salloum F, Das A, Hyder H, Kukreja RC (2005) Phosphodiesterase-5 inhibition with sildenafil attenuates cardiomyocyte apoptosis and left ventricular dysfunction in a chronic model of doxorubicin cardiotoxicity. Circulation 111:1601–1610
Greco S, Fasanaro P, Castelvecchio S, D’Alessandra Y, Arcelli D, Di Donato M, Malavazos A, Capogrossi MC, Menicanti L, Martelli F (2012) MicroRNA dysregulation in diabetic ischemic heart failure patients. Diabetes 61:1633–1641
Han H, Qu G, Han C, Wang Y, Sun T, Li F, Wang J, Luo S (2015) MiR-34a, miR-21 and miR-23a as potential biomarkers for coronary artery disease: a pilot microarray study and confirmation in a 32 patient cohort. Exp Mol Med 47:e138
Herman EH, Lipshultz SE, Rifai N, Zhang J, Papoian T, Yu ZX, Takeda K, Ferrans VJ (1998) Use of cardiac troponin T levels as an indicator of doxorubicin-induced cardiotoxicity. Cancer Res 58:195–197
Herman EH, Zhang J, Lipshultz SE, Rifai N, Chadwick D, Takeda K, Yu ZX, Ferrans VJ (1999) Correlation between serum levels of cardiac troponin-T and the severity of the chronic cardiomyopathy induced by doxorubicin. J Clin Oncol 17:2237–2243
Holmgren G, Synnergren J, Bogestal Y, Ameen C, Akesson K, Holmgren S, Lindahl A, Sartipy P (2015) Identification of novel biomarkers for doxorubicin-induced toxicity in human cardiomyocytes derived from pluripotent stem cells. Toxicology 328:102–111
Hrelia S, Fiorentini D, Maraldi T, Angeloni C, Bordoni A, Biagi PL, Hakim G (2002) Doxorubicin induces early lipid peroxidation associated with changes in glucose transport in cultured cardiomyocytes. BBA Biomembr 1567:150–156
Hsu A, Chen SJ, Chang YS, Chen HC, Chu PH (2014) Systemic approach to identify serum microRNAs as potential biomarkers for acute myocardial infarction. Biomed Res Int 418628
Jaenke RS (1974) An anthracycline antibiotic-induced cardiomyopathy in rabbits. Lab Invest 30:292–304
Kawano H, Okada R, Kawano Y, Sueyoshi N, Shirai T (1994) Apoptosis in acute and chronic myocarditis. Jpn Heart J 35:745–750
Krishnan K, Steptoe AL, Martin HC, Wani S, Nones K, Waddell N, Mariasegaram M, Simpson PT, Lakhani SR, Gabrielli B, Vlassov A, Cloonan N, Grimmond SM (2013) MicroRNA-182-5p targets a network of genes involved in DNA repair. RNA 19:230–242
Kwak HB (2013) Effects of aging and exercise training on apoptosis in the heart. J Exerc Rehabil 9:212–219
Latronico MVG, Condorelli G (2009) MicroRNAs and cardiac pathology. Nat Rev Cardiol 6:418–429
L’Ecuyer T, Sanjeev S, Thomas R, Novak R, Das L, Campbell W, Vander Heide R (2006) DNA damage is an early event in doxorubicin-induced cardiac myocyte death. Am J Physiol Heart Circ Physiol 291:H1273–H1280
Leptidis S, el Azzouzi H, Lok SI, de Weger R, Olieslagers S, Kisters N, Silva GJ, Heymans S, Cuppen E, Berezikov E, De Windt LJ, Martins PD (2013) A deep sequencing approach to uncover the miRNOME in the human heart. PLoS ONE 8(2):e57800. doi:10.1371/journal.pone.0057800
Lin RCY, Weeks KL, Gao XM, Williams RBH, Bernardo BC, Kiriazis H, Matthews VB, Woodcock EA, Bouwman RD, Mollica JP, Speirs HJ, Dawes IW, Daly RJ, Shioi T, Izumo S, Febbraio MA, Du XJ, McMullen JR (2010) PI3K(p110 alpha) protects against myocardial infarction-induced heart failure identification of PI3K-regulated miRNA and mRNA. Arterioscler Thromb Vasc Biol 30:724–732
Lipshultz SE, Miller TL, Scully RE, Lipsitz SR, Rifai N, Silverman LB, Colan SD, Neuberg DS, Dahlberg SE, Henkel JM, Asselin BL, Athale UH, Clavell LA, Laverdiere C, Michon B, Schorin MA, Sallan SE (2012) Changes in cardiac biomarkers during doxorubicin treatment of pediatric patients with high-risk acute lymphoblastic leukemia: associations with long-term echocardiographic outcomes. J Clin Oncol 30:1042–1049
Matsumoto S, Sakata Y, Suna S, Nakatani D, Usami M, Hara M, Kitamura T, Hamasaki T, Nanto S, Kawahara Y, Komuro I (2013) Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction short communication. Circ Res 113:322–326
Moskwa P, Buffa FM, Pan YF, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM, Gorospe M, Harris AL, Helleday T, Chowdhury D (2011) miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell 41:210–220
O’Brien PJ (2006) Blood cardiac troponin in toxic myocardial injury: archetype of a translational safety biomarker. Expert Rev Mol Diagn 6:685–702
O’Brien PJ, Smith DEC, Knechtel TJ, Marchak MA, Pruimboom-Brees I, Brees DJ, Spratt DP, Archer FJ, Butler P, Potter AN, Provost JP, Richard J, Snyder PA, Reagan WJ (2006) Cardiac troponin I is a sensitive, specific biomarker of cardiac injury in laboratory animals. Lab Anim 40:153–171
Olivetti G, Abbi R, Quaini F, Kajstura J, Cheng W, Nitahara JA, Quaini E, DiLoreto C, Beltrami CA, Krajewski S, Reed JC, Anversa P (1997) Apoptosis in the failing human heart. N Engl J Med 336:1131–1141
Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R, Reinhardt F, Liao R, Krieger M, Jaenisch R, Lodish HF, Blelloch R (2009) Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res 105:585–594
Saraste A, Pulkki K, Kallajoki M, Henriksen K, Parvinen M, VoipioPulkki LM (1997) Apoptosis in human acute myocardial infarction. Circulation 95:320–323
Shahzadi A, Sonmez I, Allahverdiyev O, Onal B, Kandaz C, Ozyazgan SO, Akkan AG, Yazici Z (2014) Cardiac troponin-I (cTnI) a biomarker of cardiac injuries induced by doxorubicin alone and in combination with ciprofloxacin, following acute and chronic dose protocol in Sprague Dawley rats. Int J Pharmacol 10:258–266
Sharov VG, Sabbah HN, Shimoyama H, Goussev AV, Lesch M, Goldstein S (1996) Evidence of cardiocyte apoptosis in myocardium of dogs with chronic heart failure. Am J Pathol 148:141–149
Simkhovich BZ, Kleinman MT, Kloner RA (2009) Particulate air pollution and coronary heart disease. Curr Opin Cardiol 24:604–609
Singal PK, Iliskovic N (1998) Doxorubicin-induced cardiomyopathy. N Engl J Med 339:900–905
Small EM, Frost RJA, Olson EN (2010a) MicroRNAs add a new dimension to cardiovascular disease. Circulation 121:1022–1032
Small EM, O’Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, Richardson JA, Olson EN (2010b) Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci USA 107:4218–4223
Smith LA, Cornelius VR, Plummer CJ, Levitt G, Verrill M, Canney P, Jones A (2010) Cardiotoxicity of anthracycline agents for the treatment of cancer: systematic review and meta-analysis of randomised controlled trials. BMC Cancer 10:337. doi:10.1186/1471-2407-10-337
Taurino C, Miller WH, McBride MW, McClure JD, Khanin R, Moreno MU, Dymott JA, Delles C, Dominiczak AF (2010) Gene expression profiling in whole blood of patients with coronary artery disease. Clin Sci 119:335–343
Tokarska-Schlattner M, Zaugg M, da Silva R, Lucchinetti E, Schaub MC, Wallimann T, Schlattner U (2005) Acute toxicity of doxorubicin on isolated perfused heart: response of kinases regulating energy supply. Am J Physiol Heart Circ Physiol 289:H37–H47
Tonomura Y, Mori Y, Torii M, Uehara T (2009) Evaluation of the usefulness of biomarkers for cardiac and skeletal myotoxicity in rats. Toxicology 266:48–54
Ueno M, Kakinuma Y, Yuhki KI, Murakoshi N, Iemitsu M, Miyauchi T, Yamaguchi I (2006) Doxorubicin induces apoptosis by activation of caspase-3 in cultured cardiomyocytes in vitro and rat cardiac ventricles in vivo. J Pharm Sci 101:151–158
Vacchi-Suzzi C, Bauer Y, Berridge BR, Bongiovanni S, Gerrish K, Hamadeh HK, Letzkus M, Lyon J, Moggs J, Paules RS, Pognan F, Staedtler F, Vidgeon-Hart MP, Grenet O, Couttet P (2012) Perturbation of microRNAs in rat heart during chronic doxorubicin treatment. PLoS ONE 7(7):e40395. doi:10.1371/journal.pone.0040395
Walker DB (2006) Serum chemical biomarkers of cardiac injury for nonclinical safety testing. Toxicol Pathol 34:94–104
Wallace KB (2003) Doxorubicin-induced cardiac mitochondrionopathy. Pharmacol Toxicol 93:105–115
Weber K, Rostert N, Bauersachs S, Wess G (2015) Serum microRNA profiles in cats with hypertrophic cardiomyopathy. Mol Cell Biochem 402:171–180
Yan DS, Dong XD, Chen XY, Yao SS, Wang LH, Wang J, Wang C, Hu DN, Qu J, Tu LL (2012) Role of microRNA-182 in posterior uveal melanoma: regulation of tumor development through MITF, BCL2 and Cyclin D2. PLoS ONE 7(7):e40967. doi:10.1371/journal.pone.0040967
Yan ZX, Wu LL, Xue K, Zhang QL, Guo Y, Romero M, Leboeuf C, Janin A, Chen SJ, Wang L, Zhao WL (2014) MicroRNA187 overexpression is related to tumor progression and determines sensitivity to bortezomib in peripheral T-cell lymphoma. Leukemia 28:880–887
Yao EL, Ventura A (2011) A new role for miR-182 in DNA repair. Mol Cell 41:135–137
Yeh ETH, Tong AT, Lenihan DJ, Yusuf SW, Swafford J, Champion C, Durand JB, Gibbs H, Zafarmand AA, Ewer MS (2004) Cardiovascular complications of cancer therapy—diagnosis, pathogenesis, and management. Circulation 109:3122–3131
Zhu XM, Wang HJ, Liu F, Chen L, Luo WJ, Su PX, Li WM, Yu LP, Yang XC, Cai J (2013) Identification of micro-RNA networks in end-stage heart failure because of dilated cardiomyopathy. J Cell Mol Med 17:1173–1187
Acknowledgments
This work was supported by the ‘Detection of endpoints and biomarkers for repeated dose toxicity using in vitro systems (DETECTIVE)’ project (FP7 Health Programme, European Commission).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Chaudhari, U., Nemade, H., Gaspar, J.A. et al. MicroRNAs as early toxicity signatures of doxorubicin in human-induced pluripotent stem cell-derived cardiomyocytes. Arch Toxicol 90, 3087–3098 (2016). https://doi.org/10.1007/s00204-016-1668-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-016-1668-0