Abstract
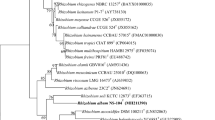
A Gram-stain negative, aerobic, non-motile, and rod-shaped novel bacterial strain, designated MAH-2T, was isolated from a soil sample of rose garden and was characterized using a polyphasic approach. The colonies were light pink color, smooth, circular and 0.2–0.6 mm in diameter when grown on nutrient agar for 3 days. Strain MAH-2T grows at 15–40 °C (optimum growth temperature 30 °C), at pH 5.0–7.0 (optimum growth pH 6.5) and at 0–2% NaCl (optimum 0-0.5%). Cell growth occurs on nutrient agar and R2A agar but not on tryptone soya agar, luria–bertani agar and MacConkey agar. The strain was positive for both catalase and oxidase tests. The strain was able to synthesis of silver nanoparticles. According to the 16S rRNA gene sequence comparisons, the isolate was identified as a member of the genus Microvirga and was most closely related to Microvirga soli R491T (96.7% sequence similarity), Microvirga subterranea Fail4T (96.4%), Microvirga guangxiensis 25BT (96.0%) and Microvirga aerophila 5420S-12T (95.9%). The genomic DNA G + C content of isolated strain was determined to be 62.5 mol% and the predominant isoprenoid quinone was Q-10. The major fatty acids were identified as summed feature 8 (comprising C18:1 ω7c and/or C18:1 ω6c) and C19:0 cyclo ω8c. On the basis of these phenotypic, genotypic, and chemotaxonomic studies and DNA–DNA hybridization results, the isolated strain MAH-2T represents a novel species, for which the name Microvirga rosea sp. nov. is proposed, with MAH-2T as the type strain (= KACC 19290T = CGMCC1.16488T).

Similar content being viewed by others
References
Ardley JK, Parker MA, de Meyer SE, Trengove RD, O’Hara GW (2012) Microvirga lupini sp. nov., Microvirga lotononidis sp. nov. and Microvirga zambiensis sp. nov. are alphaproteobacterial root-nodule bacteria that specifically nodulate and fix nitrogen with geographically and taxonomically separate legume hosts. Int J Syst Evol Microbiol 62:2579–2588
Christensen WB (1946) Urea decomposition as a means of differentiating proteus and paracolon cultures from each other and from Salmonella and Shigella types. J Bacteriol 52:461–466
Collins MD (1985) Isoprenoid quinone analyses in bacterial classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 267–287
Collins MD, Jones D (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol Rev 45:316–354
Dahal RH, Kim J (2017) Microvirga soli sp. nov., an alphaproteobacterium isolated from soil. Int J Syst Evol Microbiol 67:127–132
Du J, Singh H, Yi TH (2017) Biosynthesis of silver nanoparticles by Novosphingobium sp. THG-C3 and their antimicrobial potential. Artif Cells Nanomed Biotechnol 45:211–217
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Fautz E, Reichenbach H (1980) A simple test for flexirubin-type pigments. FEMS Microbiol Lett 8:87–91
Felsenstein J (1985) Confidence limit on phylogenies: an approach using the bootstrap. Evolution/evolution. Int J Org Evol 39:783–791
Garrity GM, Bell JA, Liburn T, Class I (2005a) Alphaproteobacteria class. nov. In: Brenner DJ, Krieg NR, Staley JT. Garrity GM (eds) Bergey’s mannual of systematic bacteriology, the proteobacteria, part C, the alpha-, beta-, delta-, and epsilonproteobacteria, vol 2, 2nd edn. Springer, New York, p 1
Garrity GM, Bell JA, Liburn T. Family IX (2005b) Methylobacteriaceae fam. nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds) Bergey’s mannual of systematic bacteriology, the proteobacteria, part C, the alpha-, beta-, delta-, and epsilonproteobacteria, vol 2, 2nd edn. Springer, New York, p 567
Gillis M, De Ley J, De Cleene M (1970) The determination of molecular weight of bacterial genome DNA from renaturation rates. Eur J Biochem 12:143–153
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Huq MA (2017) Chryseobacterium chungangensis sp. nov., a bacterium isolated from soil of sweet gourd garden. Arch Microbiol. https://doi.org/10.1007/s00203-017-1469-8
Kanso S, Patel BKC (2003) Microvirga subterranea gen. nov., sp. nov., a moderate thermophile from a deep subsurface Australian thermal aquifer. Int J Syst Evol Microbiol 53:401–406
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
McConaughy BL, Laird CD, McCarthy BJ (1969) Nucleic acid reassociation in formamide. Biochemistry 8:3289–3295
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G + C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, O’Donnel AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parleet JH (1984) An intergrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Meth 2:233–241
Moore DD, Dowhan D (1995) Preparation and analysis of DNA. In: Ausubel FW, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) Current protocols in molecular biology. Wiley, New York, pp 2–11
Radl V, Simões-Araújo JL, Leite J, Passos SR, Martins LM (2014) Microvirga vignae sp. nov., a root nodule symbiotic bacterium isolated from cowpea grown in semi-arid Brazil. Int J Syst Evol Microbiol 64:725–730
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Bio Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. Newark, DE: MIDI Inc
Skerman VBD (1967) A guide to the identification of the genera of bacteria, 2nd edn. Williams and Wilkins, Baltimore
Stabili L, Gravili C, Tredici SM, Piraino S, Talà A, Boero F, Alifano P (2008) Epibiotic Vibrio luminous bacteria isolated from some hydrozoa and bryozoa species. Microb Ecol 56:625–636
Stackebrandt E, Murray RGE, Trüper HG (1988) Proteobacteria classis nov., a name for the phylogenetic taxon that includes the ‘purple bacteria and their relatives’. Int J Syst Bacteriol 38:321–325
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Weon HY, Kwon SW, Son JA, Jo EH, Kim SJ (2010) Description of Microvirga aerophila sp. nov. and Microvirga aerilata sp. nov., isolated from air, reclassification of Balneimonas flocculans Takeda et al. 2004 as Microvirga flocculans comb. nov. and emended description of the genus Microvirga. Int J Syst Evol Microbiol 60:2596–2600
Zhang J, Song F, Xin YH, Zhang J, Fang C (2009) Microvirga guangxiensis sp. nov., a novel alphaproteobacterium from soil, and emended description of the genus Microvirga. Int J Syst Evol Microbiol 59:1997–2001
Acknowledgements
This study was performed with the support of the National Research Foundation (NRF) of Korea grant (Project no. NRF-2018R1C1B5041386) funded by Korean government, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
The NCBI GenBank accession number for the 16S rRNA gene sequence of strains MAH-2T is KY964282 and the digital protologue database (DPD) Taxon Number is TA00610.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huq, M.A. Microvirga rosea sp. nov.: a nanoparticle producing bacterium isolated from soil of rose garden. Arch Microbiol 200, 1439–1445 (2018). https://doi.org/10.1007/s00203-018-1558-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-018-1558-3