Abstract
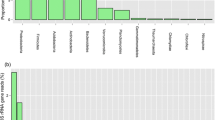
Drass is the coldest inhabited place in India and the second coldest, inhabited place in the world, after Siberia. Using the 16SrDNA amplicon pyrosequencing, bacterial diversity patterns were cataloged across the Drass cold desert. In order to identify the ecotype abundance across cold desert environment, bacterial diversity patterns of Drass were further compared with the bacterial diversity of two other cold deserts, i.e., Antarctic and Arctic. Acidobacteria, Proteobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria and Gemmatimonadetes were among the highly abundant taxonomic groups present across all the three cold deserts and were designated as the core phyla. However, Firmicutes, Nitrospirae, Armatimonadetes (former candidate division OP10), Planctomycetes, TM7, Chloroflexi, Deinococcus-Thermus, Tenericutes and candidate phyla WS3 were identified as rare phyla in Drass, Antarctic and Arctic samples. Differential abundance patterns were also computed across all the three samples, i.e., Acidobacteria (32.1 %) were dominant in Drass and Firmicutes (52.9 ± 17.6 %) and Proteobacteria (42 ± 1.3 %) were dominant in Antarctic and Arctic reference samples, respectively. Alpha diversity values Shannon’s (H) and Simpson’s (1-D) diversity indices were highest for Antarctic samples, whereas richness estimators (ACE and Chao1) were maximum for Drass soil suggesting greater species richness in bacterial communities in Drass than the Antarctic and Arctic samples.



Similar content being viewed by others
References
Adams BJ, Bardgett RD, Ayres E, Wall DH, Aislabie J, Bamforth S, Bargagli R, Cary C, Cavacini P, Connell L, Convey P, Fell JW, Frati F, Hogg ID, Newsham KK (2006) Diversity and distribution Victoria Land biota. Soil Biol Biochem 38:3003–3018
Aislabie J, Jordan S, Ayton J, Klassen JL, Barker GM, Turner S (2009) Bacterial diversity associated with ornithogenic soil of the Ross Sea region, Antarctica. Can J Microbiol 55:21–36
Amorim JH, Macena TNS, Lacerda GV (2008) An improved extraction protocol for metagenomic DNA from a soil of the Brazilian Atlantic Rainforest. Genet Mol Res 7(4):1226–1232
Bonilla S, Villeneuve V, Vincent WF (2005) Benthic and planktonic algal communities in a high arctic lake: pigment structure and contrasting responses to nutrient enrichment. J Phycol 41:1120–1130
Bottos EM, Scarrow JW, Archer SD, McDonald IR, Cary SC (2014) Bacterial community structures of Antarctic soils. In: Cowan DA (ed) Antarctic terrestrial microbiology, vol 1. Springer, Berlin, pp 9–33
Brady SF (2007) Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nat Protoc 2(5):1297–1305
Campbell BJ, Polson SW, Hanson TE, Mack MC, Schuur EAG (2010) The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ Microbiol 12(7):1842–1854
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Charan G, Bharti VK, Jadhav SE, Kumar S, Acharya S, Kumar P, Gogoi D, Srivastava RB (2013) Altitudinal variations in soil physico-chemical properties at cold desert high altitude. J Soil Sci Plant Nutr 13(2):267–277
Cheng SM, Foght JM (2007) Cultivation-independent and –dependent characterization of bacteria resident beneath John Evans Glacier. FEMS Microbiol Ecol 59:318–330
Chu H, Fierer N, Lauber CL, Caporaso JG, Knight R, Grogan P (2010) Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol 12:2998–3006
Cole JR, Wang Q, Cardenas E, Fish J, Chai B (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:141–145
Delmont TO, Robe P, Cecillon S, Clark IM, Constancias F, Simonet P, Hirsch PR, Vogel TM (2011) Accessing the soil metagenome for studies of microbial diversity. Appl Environ Microbiol 77(4):1315–1324
Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T (2008) Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 8:125
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200
Egge E, Bittner L, Andersen T, Audic S, de Vargas C, Edvardsen B (2013) 454 pyrosequencing to describe microbial eukaryotic community composition, diversity and relative abundance: a test for marine haptophytes. PLoS ONE 8(9):e74371
Fan L, McElroy K, Thomas T (2012) Reconstruction of ribosomal RNA genes from metagenomic data. PLoS ONE 7(6):e39948
Fazi S, Amalfitano S, Pernthaler J, Puddu A (2005) Bacterial communities associated with benthic organic matter in headwater stream microhabitats. Environ Microbiol 7:1633–1640
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fierer N, Schimel J, Holden P (2003) Variations in microbial community composition through two soil depth profiles. Soil Biol Biochem 35:167–176
Fierer N, Bradford M, Jackson R (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Gontcharova V, Youn E, Wolcott RD, Hollister EB, Gentry TJ (2010) Black box chimera check (B2C2): a windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene datasets. Open Microbiol J 4:47–52
Inceoglu O, Hoogwout EF, Hill P, Elsas JDV (2010) Effect of DNA extraction method on the apparent microbial diversity of soil. Appl Environ Microbiol 76(10):3378–3382
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N (2009) A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 3:442–453
Kastovska K, Elster J, Stibal M, Santruckova H (2005) Microbial assemblages in soil microbial succession after glacier retreat in Svalbard (high arctic). Microbiol Ecol 50:396–407
Kaur G, Sharma R, Singh K, Sharma P (2015) Delineating bacterial community structure of polluted soil samples collected from cancer prone belt of Punjab, India. 3 Biotech. doi:10.1007/s13205-014-0270-5
Keki Z, Grebner K, Bohus V, Marialigeti K, Toth EM (2013) Application of special oligotrophic media for cultivation of bacterial communities originated from ultrapure water. Acta Microbiol Immunol Hung 60(3):345–357
Kim HM, Jung JY, Yergeau E, Hwang CY, Hinzman L, Nam S, Hong SG, Kim OS, Chun J, Lee YK (2014) Bacterial community structure and soil properties of a subarctic tundra soil in Council, Alaska. FEMS Microbiol Ecol 89:465–475
Kirchman DL, Cottrell MT, Lovejoy C (2010) The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol 12:1132–1143
Kistler JO, Booth V, Bradshaw DJ, Wade WG (2013) Bacterial community development in experimental gingivitis. PLoS ONE 8(8):e71227
Krober M, Bekel T, Diaz NN, Goesmann A, Jaenicke S (2009) Phylogenetic characterization of a biogas plant microbial community integrating clone library 16S-rDNA sequences and metagenome sequence data obtained by 454-pyrosequencing. J Biotechnol 142:38–49
Lee CK, Barbier BA, Bottos EM, McDonald IR, Cary SC (2012) The inter-valley soil comparative survey: the ecology of Dry Valley edaphic microbial communities. ISME J 6:1046–1057
Li XL, Liu XH, Ju YT, Huang FX (2003) Properties of soils in Grove Mountains, East Antarctica. Sci China (D) 46(7):683–693
Li Y, Li F, Zhang X, Qin S, Zeng Z, Dang H, Qin Y (2008) Vertical distribution of bacterial and archaeal communities along discrete layers of a deep-sea cold sediment sample at the East Pacific Rise (13°N). Extremophiles 12:573–585
Liles MR, Williamson LL, Rodbumrer J, Torsvik V, Parsley LC, Goodman RM, Handelsman J (2009) Isolation and cloning of high-molecular-weight metagenomic DNA from soil microorganisms. Cold Spring Harb Protoc 8:pdb.prot5271R
Liu Y, Yao T, Jiao N, Kang S, Xu B, Zeng Y, Huang S, Liu X (2009) Bacterial diversity in the snow over Tibetan Plateau Glaciers. Extremophiles 13:411–423
Moller AK, Soborg DA, Abu Al-Soud W, Sorensen SJ, Kroer N (2013) Bacterial community structure in High-Arctic snow and freshwater as revealed by pyrosequencing of 16rRNA genes and cultivation. Polar Res 32:17390
Naether A, Foesel BU, Naegele V, Wust PK, Weinert J, Bonkowski M, Alt F, Oelmann Y, Polle A, Lohaus G, Gockel S, Hemp A, Kalko EKV, Linsenmair KE, Pfeiffer S, Renner S, Schoning I, Weisser WW, Wells K, Fischer M, Overmann J, Friedrich MW (2012) Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl Environ Microbiol 78(20):7398–7406
Namgail T (2009) Mountain ungulates of the Trans-Himalayan region of Ladakh, India. Int J Wilder 15:35–40
Niederberger TD, Sohm JA, Gunderson TE, Parker AE, Tirindelli J, Capone DG, Carpenter EJ, Cary SC (2015) Microbial community composition of transiently wetted Antarctic Dry Valley soils. Front Microbiol 6(9). doi:10.3389/fmicb.2015.00009
Nossa CW, Oberdorf WE, Yang L, Aas JA, Paster BJ, Desantis TZ, Brodie EL, Malamud D, Poles MA, Pei Z (2010) Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J Gastroenterol 16(33):4135–4144
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2013) Vegan: community ecology package. R package version 2.0-7
Pang M, Abdullah N, Lee C, Ng CC (2008) Isolation of high molecular weight DNA from forest topsoil for metagenomic analysis. Asia Pac J Mol Biol Biotechnol 16:35–41
Pointing SB, Chan Y, Lacap DC, Lau MCY, Jurgens JA, Farrell RL (2009) Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci USA 106:19964–19969
Pradhan S, Srinivas TN, Pindi PK, Kishore KH, Begum Z, Singh PK, Singh AK, Pratibha MS, Yasala AK, Reddy GS, Shivaji S (2010) Bacterial biodiversity from Roopkund Glacier, Himalayan mountain ranges, India. Extremophiles 14:377–395
Price MN, Dehal PS, Arkin AP (2010) FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5(3):e9490
Rawat GS, Adhikari BS (2005) Floristics and distribution of plant communities across moisture and topographic gradients in Tso Kar basin, Changthang plateau, eastern Ladakh. Arct Antarct Alp Res 37:539–544
Rhoads DD, Wolcott RD, Sun Y, Dowd SE (2012) Comparison of culture and molecular identification of bacteria in chronic wounds. Int J Mol Sci 13:2535–2550
Rosenberg MS, Anderson CD (2011) PASSaGE: pattern analysis, spatial statistics and geographic exegesis. version 2. Methods Ecol Evol 2:229–232
Sagwal SS (1997-12) Ladakh: ecology and environment. APH Publishing Corporation, India
Schutte UME, Abdo Z, Foster J, Ravel J, Bunge J, Solheim B, Forney LJ (2010) Bacterial diversity in a glacier foreland of the high Arctic. Mol Ecol 19:54–66
Serkebaeva YM, Kim Y, Liesack W, Dedysh SN (2013) Pyrosequencing-based assessment of the bacteria diversity in surface and subsurface peat layers of a Northern Wetland, with focus on poorly studied phyla and candidate divisions. PLoS ONE 8(5):e63994
Shimodaira H, Hasegawa M (2001) CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246–1247
Shivaji S, Reddy GSN, Aduri RP, Kutty R, Ravenschlag K (2004) Bacterial diversity of a soil sample from Schirmacher Oasis, Antarctica. Cell Mol Biol 50:525–536
Shivaji S, Pratibha MS, Sailaja B, Hara Kishore K, Singh AK, Begum Z, Anarasi U, Prabagaran SR, Reddy GS, Srinivas TN (2011) Bacterial diversity of soil in the vicinity of Pindari glacier, Himalayan mountain ranges, India, using culturable bacteria and soil 16S rRNA gene clones. Extremophiles 15:1–22
Skidmore M, Anderson SP, Sharp M, Foght J, Lanoil BD (2005) Comparison of microbial community composition in two subglacial environments reveals a possible role for microbes in chemical weathering processes. Appl Environ Microbiol 71:6986–6997
Srinivas TNR, Singh SM, Pradhan S, Pratibha MS, Kishore KH, Singh AK, Begum Z, Prabagaran SR, Reddy GSN, Shivaji S (2011) Comparison of bacterial diversity in proglacial soil from Kafni Glacier, Himalayan Mountain ranges, India, with the bacterial diversity of other glaciers in the world. Extremophiles 15:673–690
Steven B, Briggs G, McKay CP, Pollard WH, Greer CW, Whyte LG (2007) Characterization of the microbial diversity in a permafrost sample from the Canadian High Arctic using culture-dependent and culture-independent methods. FEMS Microbiol Ecol 59(2):513–523
Stres B, Sul WJ, Murovec B, Tiedje JM (2014) Recently deglaciated high-altitude soils of the Himalaya: diverse environments, heterogenous bacterial communities and long-range dust inputs from the upper troposphere. PLoS ONE 8(9):e76440
Suzuki R, Shimodaira H (2006) Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22(12):1540–1542
Tytgat B, Verleyen E, Obbels D, Peeters K, De Wever A (2014) Bacterial diversity assessment in Antarctic terrestrial and aquatic microbial mats: a comparison between bidirectional pyrosequencing and cultivation. PLoS ONE 9(6):e97564
Varin T, Lovejoy C, Jungblut AD, Vincent WF, Corbeil J (2012) Metagenomic analysis of stress genes in microbial mat communities from Antarctica and the high Arctic. Appl Environ Microbiol 78(2):549–559
Vaz-Moreira I, Egas C, Nunes OC, Manaia CM (2011) Culture-dependent and culture-independent diversity surveys target different bacteria: a case study in a freshwater sample. Antonie Van Leeuwenhoek 100(2):245–257
Vincent WF (2007) Cold tolerance in cyanobacteria and life in the cryosphere. In: Seckbach J (ed) Algae and cyanobacteria in extreme environments. Springer, Heidelberg, pp 287–301
Wagner D, Kobabe S, Liebner S (2009) Bacterial community structure and carbon turnover in permafrost-affected soils of the Lena Delta, northeastern Siberia. Can J Microbiol 55:77–83
Wechter P, Williamson J, Robertson A, Kluepfel D (2003) A rapid, cost-effective procedure for the extraction of microbial DNA from soil. World J Microbiol Biotechnol 19:85–91
Wu X, Xi W, Ye W (2007) Bacterial community composition of a shallow hypertrophic freshwater lake in China, revealed by 16S rRNA gene sequences. FEMS Microbiol Ecol 61:85–96
Wu X, Zhang W, Liu G, Yang X, Hu P, Chen T, Zhang G, Li Z (2012) Bacterial diversity in the foreland of the Tianshan No. 1 glacier, China. Environ Res Lett 7:014038
Xiang SR, Yao TD, An LZ, Xu BQ, Li Z, Wu GJ, Wang YQ, Ma S, Chen XR (2004) Bacterial diversity in Malan ice core from the Tibetan Plateau. Folia Microbiol 49:269–275
Xuezheng L, Zhen W, Shuai C, Song W, Dan YI (2014) Bacterial diversity in Arctic marine sediment determined by culture-dependent and -independent approaches. Adv Polar Sci 25(1):46–53
Zakhia F, Jungblut AD, Taton A, Vincent WF, Wilmotte A (2008) Cyanobacteria in cold environments. In: Margesin R, Schinner F, Marx JC, Gerday C (eds) Psychrophiles: from biodiversity to biotechnology. Springer, Heidelberg, pp 121–135
Zeglin LH, Dahm CN, Barrett JE, Gooseff MN, Fitpatrick SK, Takacs-Vesbach CD (2011) Bacterial community structure along moisture gradients in the parafluvial sediments of two ephemeral desert streams. Microbiol Ecol 61:543–556
Zhou J, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Zhou J, Davey ME, Figueras JB, Rivkina E, Gilichinsky D, Tiedje JM (1997) Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 143:3913–3919
Acknowledgments
Authors are thankful to DBT Government of India (BT/PR11727/BCE/08/720/2008) for funding of this project. PG is thankful to CSIR (9/100/0177) 2K13-EMR-I, Government of India for CSIR-SRF. Ms. Sneha Ganjoo, project fellow in the school, is acknowledged for her suggestions in the preparation of Manuscript. Resources used in the Bioinformatic Centre funded by DBT Government of India are also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Rights and permissions
About this article
Cite this article
Gupta, P., Sangwan, N., Lal, R. et al. Bacterial diversity of Drass, cold desert in Western Himalaya, and its comparison with Antarctic and Arctic. Arch Microbiol 197, 851–860 (2015). https://doi.org/10.1007/s00203-015-1121-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-015-1121-4